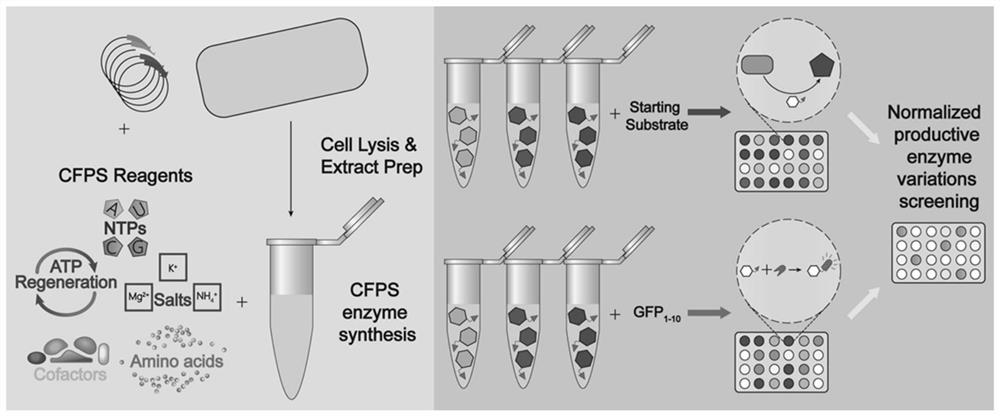
Method for quantitatively detecting target protein yield of cell-free protein synthesis system and method for screening zymoprotein with high catalytic activity
A technology for protein synthesis and quantitative detection, applied in the field of molecular biology and synthetic biology, can solve the time-consuming and labor-intensive problems of expression and purification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
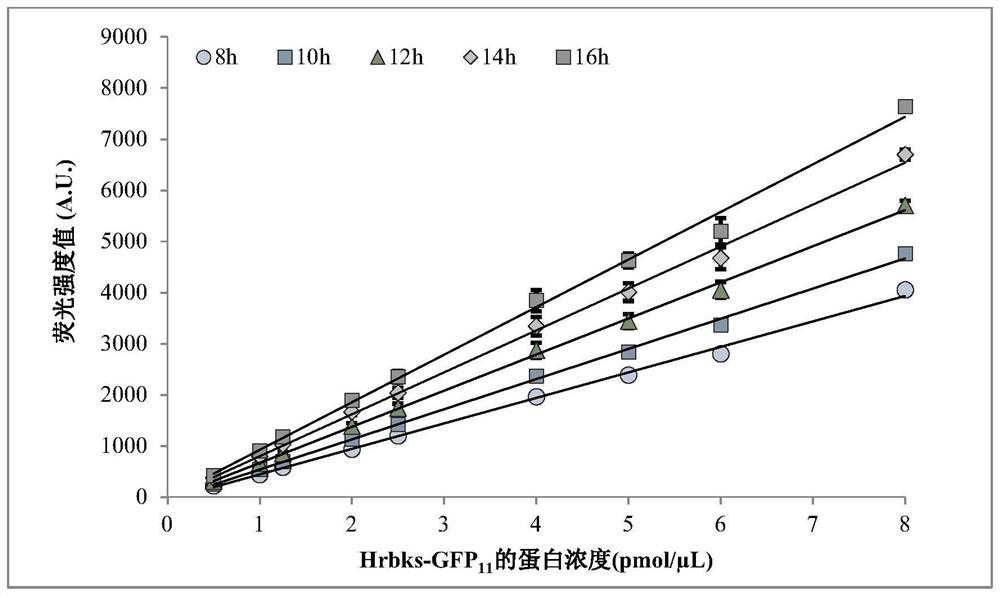
[0093] Embodiment 1 expresses Hrbks-GFP 11 Construction of fusion protein plasmid vector and Hrbks-GFP 11 Establishment of the standard curve between fluorescence intensity value and protein concentration of fusion protein under different incubation time conditions
[0094] (1) Connect the GFP11 gene to the C-terminus of the human (Homo sapiens) ribokinase HRBKS gene (NCBI: NP_071411.1) (see reference "Cabantous, Stéphanie, Waldo, et al. In vivo and in vitroprotein solubility assays using split GFP .[J].Nature Methods,2006." GFP 11M3 OPT , referred to as GFP11 in the embodiments, its nucleotide sequence is shown in SEQ ID NO.1: 5'-CGTGACCACATGGTCCTTCATGAGTACGTAAATGCTGCTGGGATTACA-3'), and in order to ensure the active expression of the HRBKS gene, a section is inserted between the HRBKS gene and the GFP11 gene Linker sequence (nucleotide sequence shown in SEQ ID NO.2: 5'-GATGGAGGGTCTGGTGGCGGATCAACAAGT-3') to obtain the complete HRBKS-GFP11 gene sequence (SEQ ID NO.4). Entru...
Embodiment 2
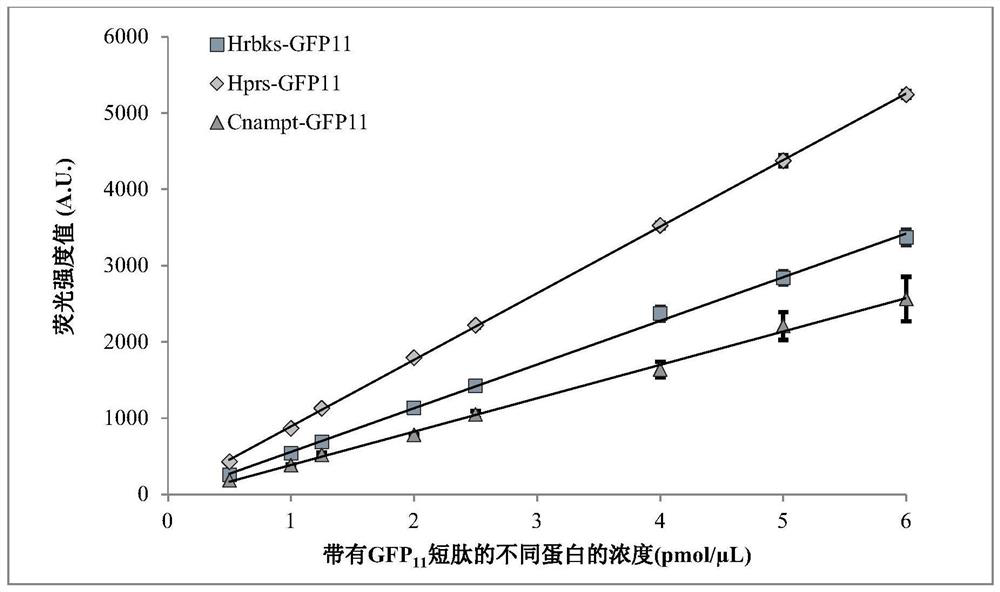
[0109] Example 2Hrbks-GFP 11 , Hprs-GFP 11 with Cnampt-GFP 11 The establishment of the standard curve between the fluorescence intensity value and the protein concentration of the fusion protein under the condition of the same incubation time
[0110] (1) Referring to step (1) plasmid pET28a-Hrbks-GFP in Example 1 11 The method of construction is to construct the human (Homo sapiens) ribose phosphate pyrophosphate kinase HPRS gene (Uniprot: P60891) and the nicotinamide phosphoribosyltransferase CNAMPT gene (GenBank : RYF34637.1) plasmid vector pET28a-Hprs-GFP 11 with pET28a-Cnampt-GFP 11 .
[0111] (2) Plasmid pET28a-Hprs-GFP 11 with pET28a-Cnampt-GFP 11 Transform into BL21 (DE3) competent cells respectively, and carry out positive transformant selection on LBK kanamycin resistance plate.
[0112] (3) Select the correct transformants identified in step (2) and refer to the literature (Li L, Liao Y, Luo Y, et al. Improved Efficiency of the Desulfurization of Oil Sulfur ...
Embodiment 3
[0115] Example 3 Using the CFPS system to express a variety of fusion proteins and detect the fluorescence intensity values under the same time conditions as the luminescence liquid incubation
[0116] (1) Referring to step (1) plasmid pET28a-Hrbks-GFP in Example 1 11 The construction method, the ribose kinase ERBKS gene (Uniprot: P0A9J6) derived from Escherichia coli, the ribose phosphate pyrophosphate kinase PPRS gene (NCBI: ABO08552.1) derived from Pyrobaculum calidifontis (NCBI: ABO08552.1), Plasmids carrying the human (Homo sapiens) nicotinamide phosphoribosyltransferase HNAMPT gene (Uniprot: P43490) and the Thermus ruber (Meiothermus ruber)-derived nicotinamide phosphoribosyltransferase MNAMPT gene (NCBI: ADD29592.1) Vector pET28a-Erbks-GFP 11 , pET28a-Pprs-GFP 11 , pET28a-Hnampt-GFP 11 with pET28a-Mnampt-GFP 11 .
[0117] (2) with pET28a-Erbks-GFP 11 , pET28a-Pprs-GFP 11 , pET28a-Hnampt-GFP 11 , pET28a-Mnampt-GFP 11 With the pET28a-Hrbks-GFP in embodiment 1 s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com