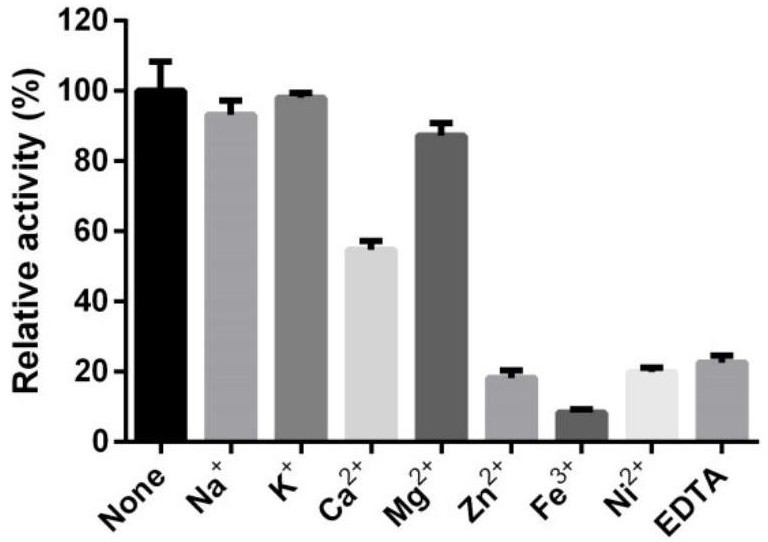
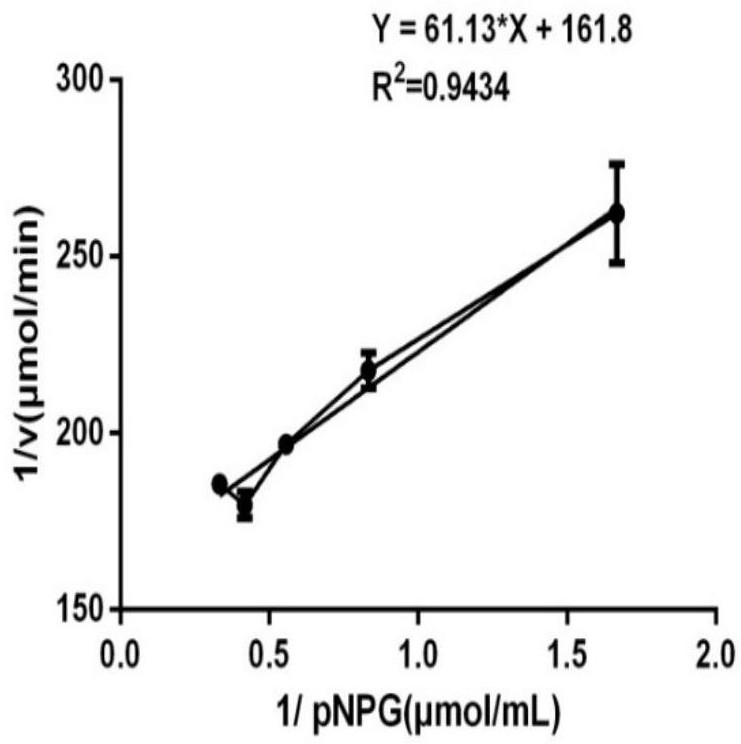
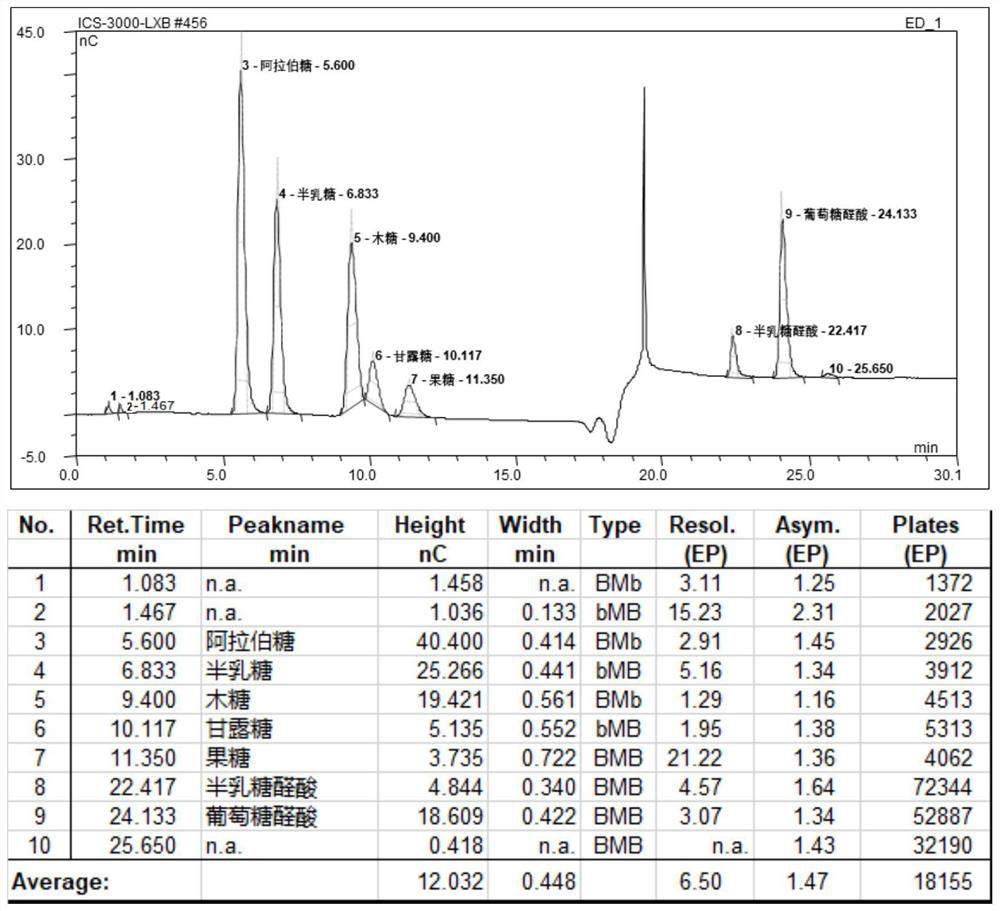
Application of arabinogalactosidase coding gene 00578 in preparation of recombinant peach gum polysaccharide hydrolase
A technology of arabinogalactose and arabinogalactan, which is applied in the field of peach gum polysaccharide lyase, can solve the problems of unsuitable commercial preparation of peach gum hydrolytic enzyme preparations and low purity, and achieve good peach gum polysaccharide degradation activity and large The effect of applying the foreground
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 Cloning of the gene orf-00578 encoding enzymes related to the degradation of peach gum polysaccharides and construction of recombinant plasmids
[0047] 1. Method
[0048] 1. Design of PCR primers
[0049] According to the ORF 00578 gene sequence obtained from Microbacterium genome sequencing, PCR primers were designed using the software Primer premier5.0, and the primer sequences are shown in Table 1 below. The primers were synthesized by Shanghai Sangon Bioengineering Company.
[0050] Table 1
[0051]
[0052] 2. PCR reaction
[0053] (1) Using Microbacterium genomic DNA as a PCR template, add each component as shown in Table 2, and mix;
[0054] Table 2
[0055]
[0056]
[0057] (2) Set the following program for PCR reaction
[0058] PCR reaction conditions: pre-denaturation at 98°C for 10 min; denaturation at 98°C for 45 sec, annealing at 55°C for 40 sec, extension at 72°C for 1 min, a total of 35 cycles; extension at 72°C for 8 min.
[0...
Embodiment 2
[0091] Example 2 Expression Analysis of Peach Gum Polysaccharide Degradation Related Enzyme Encoding Gene orf-00578 Recombinant Plasmid
[0092] 1. The growth curve of recombinant Escherichia coli BL21 was measured, and the logarithmic growth phase of Escherichia coli BL21 (DE3) was measured between 2 and 2.5 hours, and the corresponding A600nm was between 0.5 and 0.8, so this time period was selected It is the starting point for subsequent IPTG induction.
[0093] 2. Induced expression of recombinant target protein
[0094] (1) Escherichia coli BL21 expression bacteria and plasmid empty bacteria were inoculated into liquid LB medium containing corresponding antibiotics (Kan) at a 1% inoculum size, and activated overnight in a constant temperature shaker at 37°C and 220 rpm.
[0095] (2) The next day, transfer the transfer bacteria solution to fresh LB medium at 1%, and cultivate until the OD of Escherichia coli is detected by the spectrophotometer 600 When the value is 0.5-...
Embodiment 3
[0129] Example 3 Verification of polysaccharide hydrolysis function of 00578 target protein
[0130] 1. Preparation of standard curve for the determination of reducing sugar by DNS colorimetric method
[0131] (1) Take 6 EP tubes and number them sequentially;
[0132] (2) Add 200 μL of standard glucose solutions with different concentrations, the concentrations are 0, 0.1 mg / mL, 0.2 mg / mL, 0.3 mg / mL, 0.4 mg / mL, 0.5 mg / mL;
[0133] (3) Add 100 μL 3,5-dinitrosalicylic acid reagent and shake well;
[0134](4) Heating in a boiling water bath for 5 minutes, taking it out immediately after the water bath to cool, and measuring the change of the absorbance value of each system at 540nm;
[0135] (5) Draw a standard curve according to the absorbance value and the concentration of the reducing sugar solution.
[0136] 2. DNS chromogenic method to determine the enzyme activity of arabinogalactosidase
[0137] Add 40 μL 6% peach gum solution and 40 μL crude enzyme solution (that is, ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Enzyme activity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com