Recombinant protein structural domain enhanced TET enzyme and whole genome DNA methylation detection method
一种重组蛋白、全基因组的技术,应用在化学仪器和方法、生物化学设备和方法、组合化学等方向,能够解决背景噪音高、准确性低、DNA损伤大等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
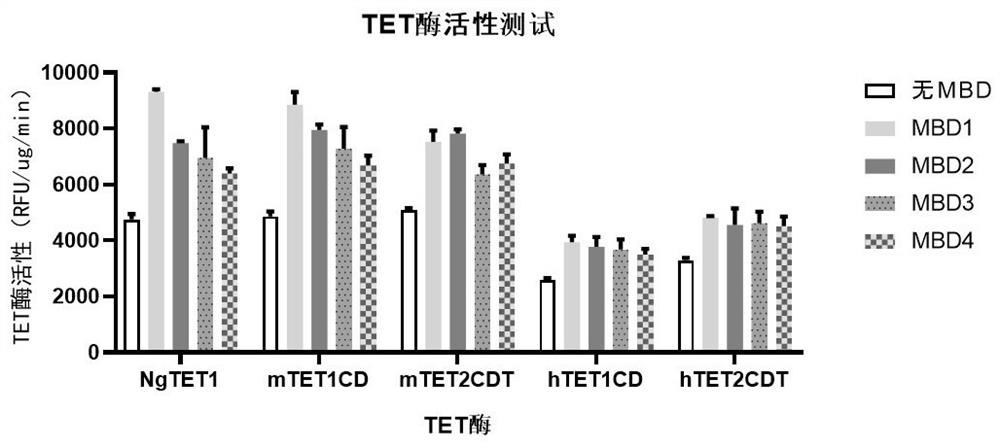
[0042] Example 1: Test of the specific activity of each recombinant protease of MBD-TET.
[0043] Epigentek's Epigenase 5mC-hydroxylase TET activity / inhibition assay kit (fluorescence method) was used to measure the enzyme specific activity of each MBD-TET recombinant protein according to the procedure in the manual.
[0044] The schematic diagram of the modified enzyme activity is shown in figure 1 , see the result figure 2 , TET enzymes fused with MBD can significantly enhance the activity of each TET enzyme.
Embodiment 2
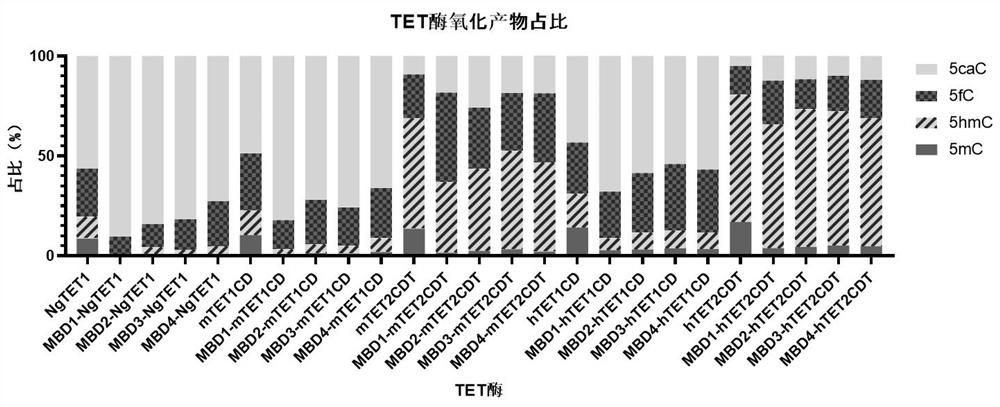
[0045] Example 2: Determination of the oxidation ability of each recombinant protease of MBD-TET to 5mC oligo.
[0046] In this embodiment, the ratio of the oxidation product of MBD-TET enzyme to 5mC is determined, and the specific implementation method is as follows:
[0047] Table 1
[0048] components Dosage 5mC oligo 1-100ng 10×TET Enzyme Reaction Buffer 3 μL 10 μM each TET enzyme 2-10 μL Add ddH 2 O to
30μL
[0049] 10×TET enzyme reaction buffer contains 1-100 mM 3-(N-morpholino)propanesulfonic acid sodium salt, 1-100 mM bis(2-hydroxyethyl)amino-tris(hydroxymethyl)methane, 1 -100 mM hydroxyethylpiperazine ethanesulfonic acid, 1-300 mM sodium chloride, 0.1-10 mM ascorbic acid, 0.1-10 mM citric acid, 0.1-20 mM α-ketoglutarate, 0.1-20 mM 1,3- Acetone dicarboxylic acid, 0.1-20 mM adenosine triphosphate, 0.1-10 mM tetrafluoro-p-benzoquinone, 0.1-10 mM tetrachloro-p-benzoquinone, 0.1-10 mM tetrabromo-p-benzoquinone, 0.1-10 mM te...
Embodiment 3
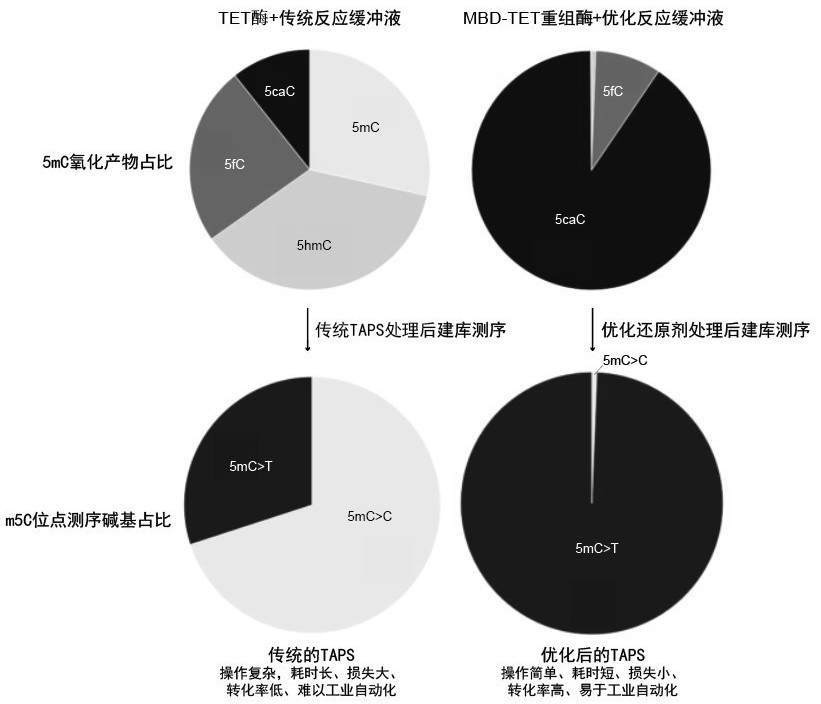
[0055] Example 3: A simple high-throughput sequencing method based on MBD1-NgTET1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com