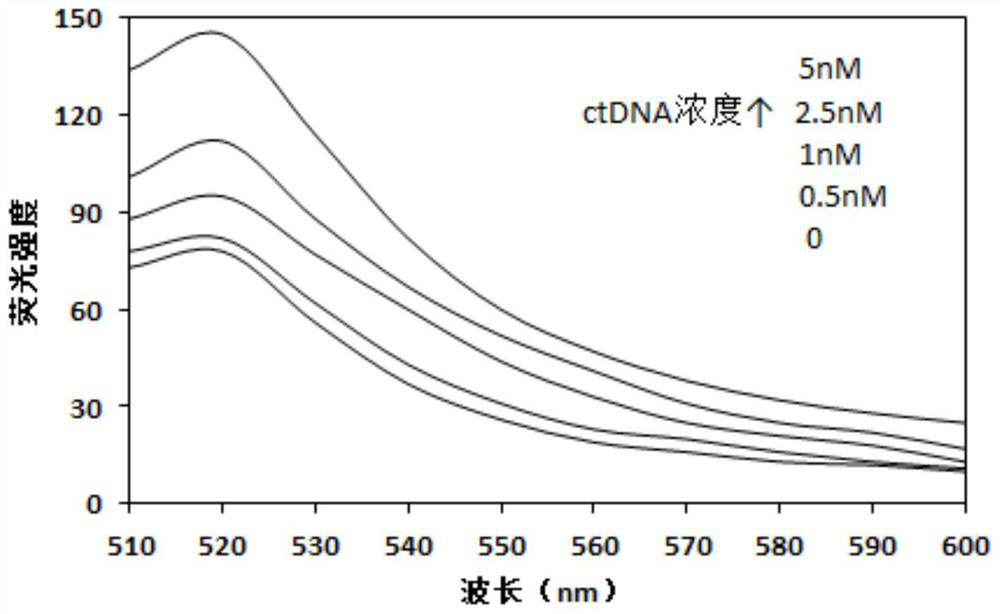
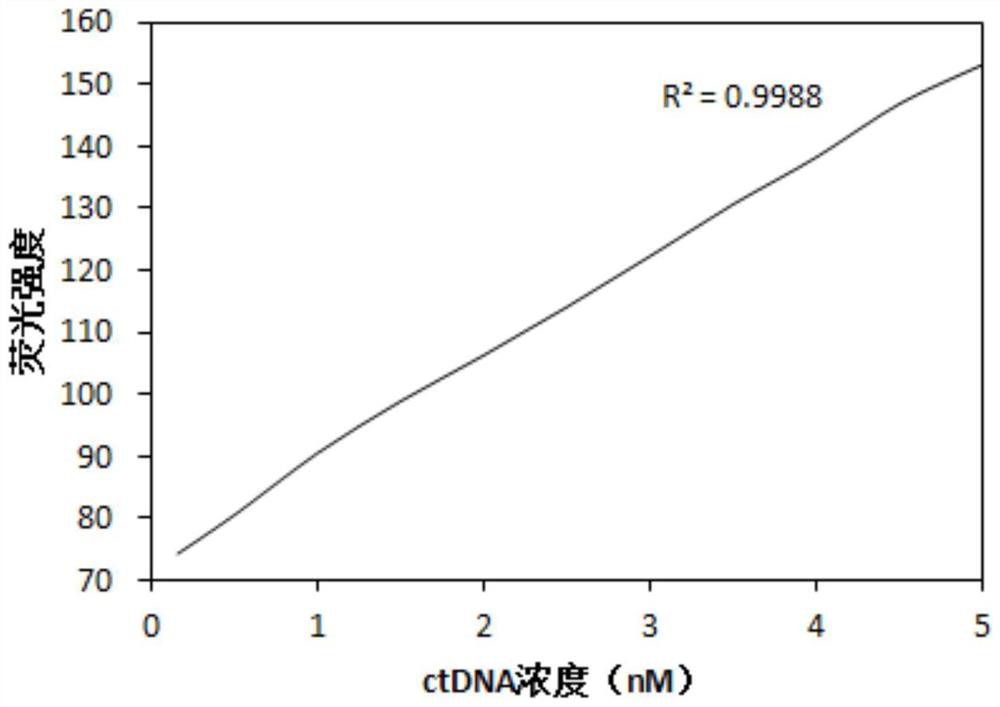
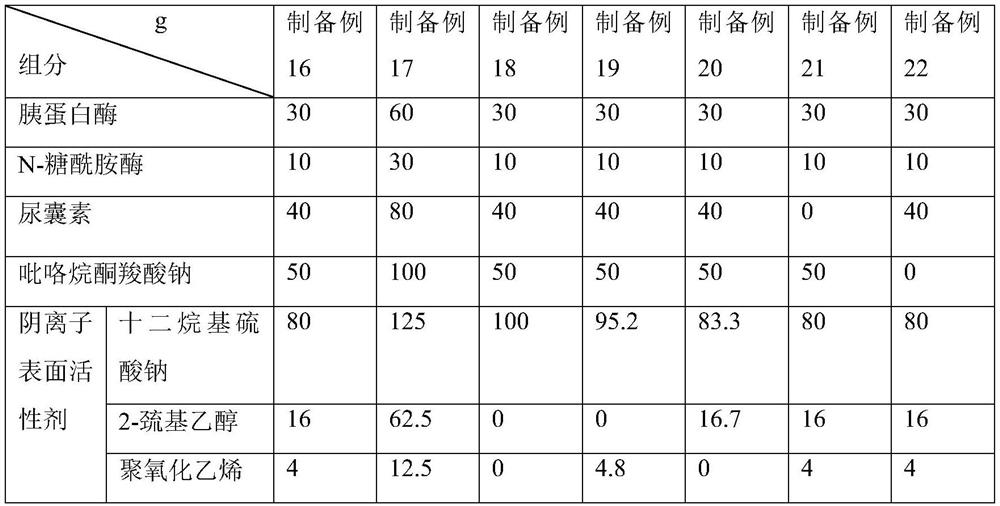
Method for optimizing ctDNA detection accuracy
An accurate, silica technology, applied in the field of tumor DNA detection, can solve the problems of high cost of gene chip detection, low detection sensitivity, non-specific signals, etc., to enhance thermal cycle stability, good dispersion, and prevent blood hanging. wall effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example 1-5
[0043] In the preparation example 1-5, the isocyanate silane coupling agent is selected from Nanjing Quanxi Chemical Co., Ltd., the article number is QX-225; the nano-lanthanum oxide is selected from Zhengzhou Chengao Chemical Products Co., Ltd., the article number is 526-98-70, and the particle size is 40nm.
preparation example 1
[0044] Preparation Example 1: Dissolve 40g of heparin sodium in 100g of sodium citrate buffer solution with a pH value of 4.5, add 40kg of 1-ethyl-3(3-dimethylpropylamine), stir well, and place at 2°C 5h, the sodium heparin solution was added to 100g concentration of chitosan in the acetic acid solution of 3%, under nitrogen protection, stirred evenly at room temperature, after centrifugation and drying, added 300g nano lanthanum oxide treated with isocyanate silane coupling agent, After mixing evenly, dry in vacuum. The method for treating nano-lanthanum oxide with isocyanate-silane coupling agent is: put 10g of isocyanate-silane coupling agent in 200g of deionized water, add 100g of nano-lanthanum oxide, ultrasonically disperse at room temperature for 20min, filter, and dry.
preparation example 2
[0045] Preparation Example 2: Dissolve 80g of heparin sodium in 200g of sodium citrate buffer solution with a pH value of 4.7, add 80g of 1-ethyl-3(3-dimethylpropylamine), stir well, and place at 6°C 5h, the sodium heparin solution was added to 200g concentration of 4% chitosan in the acetic acid solution, under nitrogen protection, stirred evenly at room temperature, after centrifugation and drying, added 300g nano lanthanum oxide treated with isocyanate silane coupling agent, After mixing evenly, dry in vacuum. The method for treating nano-lanthanum oxide with isocyanate-silane coupling agent is: put 10g of isocyanate-silane coupling agent in 200g of deionized water, add 100g of nano-lanthanum oxide, ultrasonically disperse at room temperature for 20min, filter, and dry.
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com