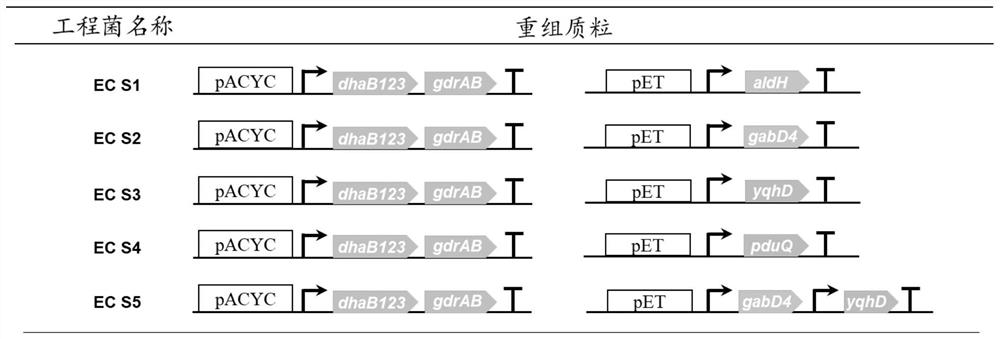
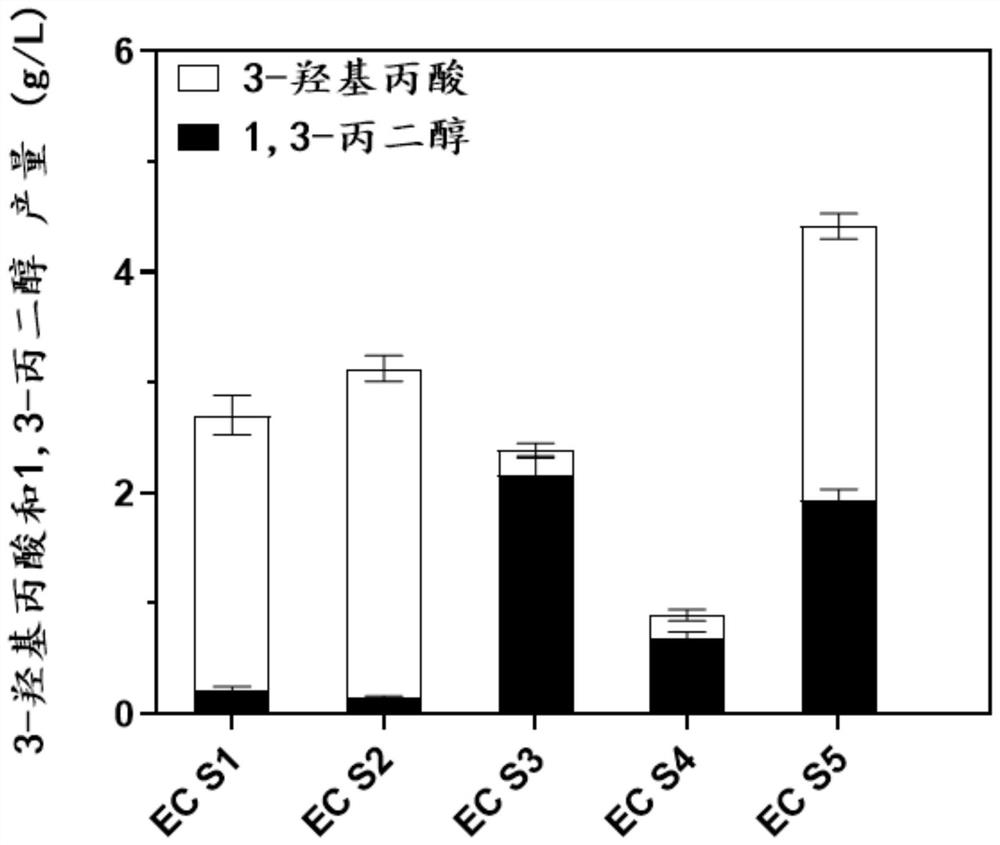
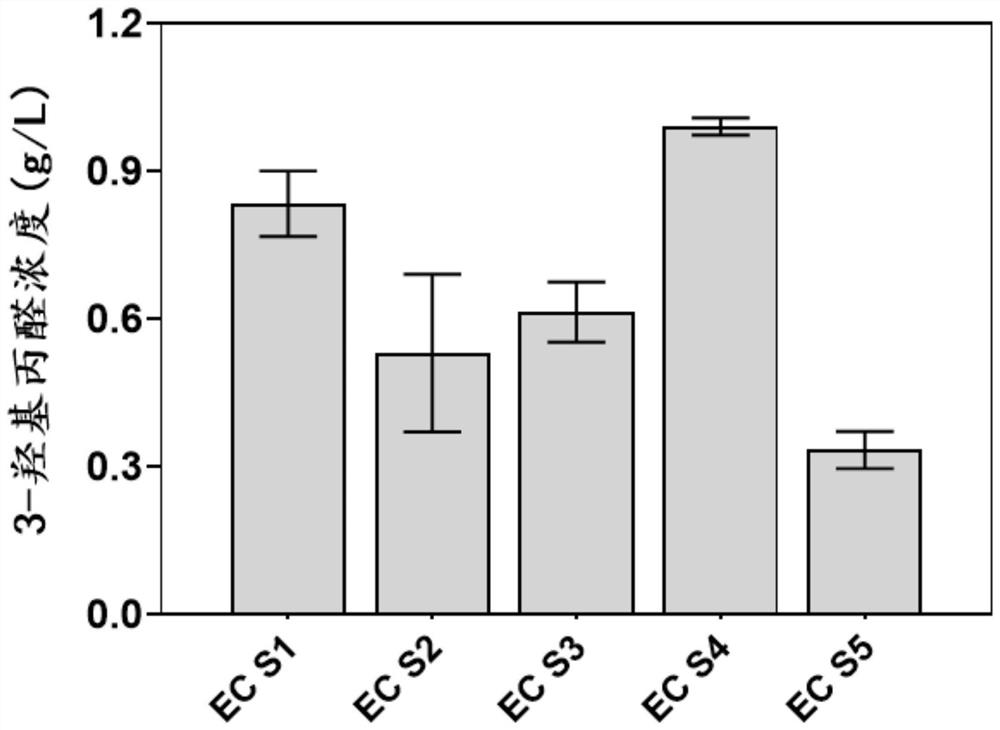
Genetically engineered bacterium EC01 S7 for co-production of 3-hydracrylic acid and 1, 3-propylene glycol as well as construction method and application of genetically engineered bacterium EC01 S7
A technology of genetically engineered bacteria and hydroxypropionic acid, applied in the field of bioengineering, can solve problems such as redox state imbalance, accumulation of toxic 3-hydroxypropanal, and insufficient activity, and achieve the effect of high-efficiency co-production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1: Construction of glycerol dehydratase and its reactivation factor (DhaB123-GdrAB) recombinant plasmid
[0042] (1) According to the Klebsiella pneumoniae glycerol dehydratase and its reactivation factor DhaB123-GdrAB gene sequence, primers were designed using Oligo7.0 software:
[0043] DhaB F: GTTTAACTTTAATAAGGAGATATACCatgaaaagatcaaaacgatttgcagtactg (SEQ ID No: 15);
[0044] GdrA R: CGGCCCCCTCGTTAACACttaattcgcctgaccggccag (SEQ ID No: 16);
[0045] GrdB F: CTGGCCGGTCAGGCGAATTAAgtgttaacgagggggccgtc (SEQ ID No: 17);
[0046] GrdB R: TTATGCGGCCGCAAGCTTGTCGACtcagtttctctcacttaacggcaggac (SEQ ID No: 18).
[0047] (2) Amplify DhaB123-GdrA gene using DhaB F and GdrA R primer pair, and amplify GdrB gene using GdrB F and GdrB R primer pair, PCR reaction parameters: pre-denaturation, 98℃ for 3min; denaturation, 98℃ for 10s; annealing, 55°C for 10s; extension, 72°C for 1.5 min; final extension, 72°C for 5 min; DhaB123-GdrA gene and GdrB gene were obtained after 33 cycl...
Embodiment 2
[0052] Embodiment 2: the construction of genetically engineered bacteria producing 3-hydroxypropionic acid or 1,3-propanediol
[0053] (1) According to the gene sequences of propionaldehyde dehydrogenase PuuC and GabD4, the gene sequences of 1,3-propanediol oxidoreductase isoenzyme YqhD, 1,3-propanediol oxidoreductase PduQ, and the sequence characteristics of the pETDuet-1 plasmid, use Oligo7.0 software designed primers. Wherein, the nucleotide sequence of PuuC is shown in SEQ ID No:3, and the amino acid sequence is shown in SEQ ID No:4; the nucleotide sequence of GabD4 is shown in SEQ ID No:5, and the amino acid sequence is shown in SEQ ID No:5. Shown in SEQ ID No:6; the nucleotide sequence of YqhD is shown in SEQ ID No:7, and the amino acid sequence is shown in SEQ ID No:8; the nucleotide sequence of PduQ is shown in SEQ ID No:9 As shown, the amino acid sequence is shown in SEQ ID No:10.
[0054] The designed primer sequences are as follows:
[0055] PuuCF F: GTTTAACTTTAA...
Embodiment 3
[0070] Example 3: Construction of genetically engineered bacteria co-producing 3-hydroxypropionic acid and 1,3-propanediol:
[0071] (1) According to the recombinant plasmid pS3 sequence and YqhD gene sequence, use Oligo7.0 software to design primers:
[0072] Backbone1 F: attagttaagtataagaaggatatacat (SEQ ID No: 27);
[0073] Backbone1 F: gtggcagcagcctaggttaa (SEQ ID No: 28);
[0074] YqhD2 F: ATTAGTTAAGTATAAGAAGGAGATATACATatgaacaactttaatctgcacac (SEQ ID No: 29);
[0075] YqhD2 R: GTGGCAGCAGCCTAGGTTAAttagcgggcggcttcgtat (SEQ ID No: 30).
[0076] (2) Using the recombinant plasmid pS2 as a template, use primer pairs Backbone1 F and Backbone1 R to amplify to obtain a linearized pS2 backbone, use YqhD2 F and YqhD2 R primer pairs to amplify the YqhD gene on the recombinant plasmid pS3, and use Gibson to assemble at 50 The YqhD gene was connected to the linearized pS2 backbone by reacting at ℃ for 30 min to obtain a recombinant plasmid that co-expressed GabD4 and YqhD.
[0077]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com