Polypeptides
A nucleic acid and cellobiose technology, applied in the field of polypeptides, can solve problems such as difficulties in industrial production of enzymes and inability to produce enzymes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] Construction of recombinant DNA containing open reading frame PH1171 in the genome of Pyrococcus horikoshii OT3.
[0088] (1) Preparation of DNA containing open reading frame PH1171.
[0089] Synthesize oligonucleotide 1171FN containing the nucleotide sequence of SEQ ID NO: 3 and oligonucleotide 1171RA containing the nucleotide sequence of SEQ ID NO: 4 according to the nucleotide sequence of the Pyrococcus horikoshi OT3 genome, so that by For PCR, a 1.6-Kb amplified DNA fragment containing the open reading frame PH1171 was obtained using the Pyrococcus horikoshii OT3 genomic DNA as a template.
[0090] Pyrococcus horikoshi OT3JCM9974 (purchased from Riken) was cultured in a medium described in JCM catalog (Riken) at 95°C for 16 hours. Purification of genomic DNA from cultured cells.
[0091] In order to obtain the DNA containing the open reading frame PH1171, oligonucleotides 1171FN and 1171RA were used as primer pairs, and the above-mentioned genomic DNA was used as ...
Embodiment 2
[0099] Production of the polypeptides of the invention.
[0100] (1) Expression of polypeptide
[0101] pECEL211 prepared in Example 1(3) or pET21d as a vector control was used to transform Escherichia coli BL21(DE3) (Novagen). Each resulting transformant was inoculated in 5 ml of LB medium containing 100 μg / ml ampicillin, respectively, and cultured overnight at 37° C. under oxygen-enriched conditions. At a concentration of 1%, the culture was inoculated into the same fresh medium and grown at 37°C under oxygen enrichment. When the turbidity reaches OD 600 =0.4-0.7, isopropyl-β-D-thiogalactopyranoside (IPTG, TakaraShuzo) was added to make the final concentration 1 mM. The cells were further incubated overnight at 15°C. After culturing, cells were collected by centrifugation, suspended in 0.5 ml of 100 mM sodium citrate buffer (pH 5.0) and disrupted by sonication. The supernatant was collected by centrifugation and used as a cell-free extract.
[0102] When cellulose is d...
Embodiment 3
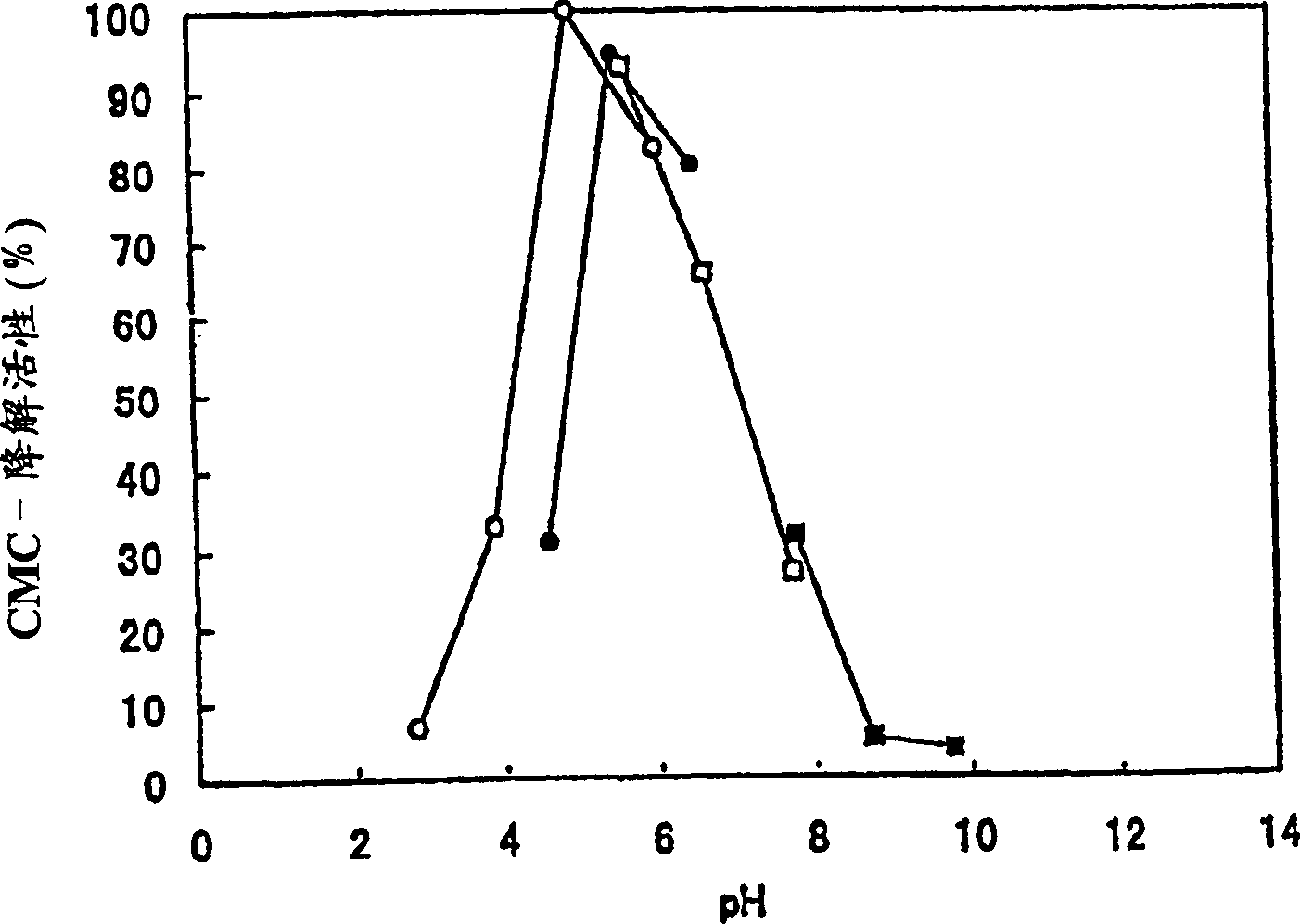
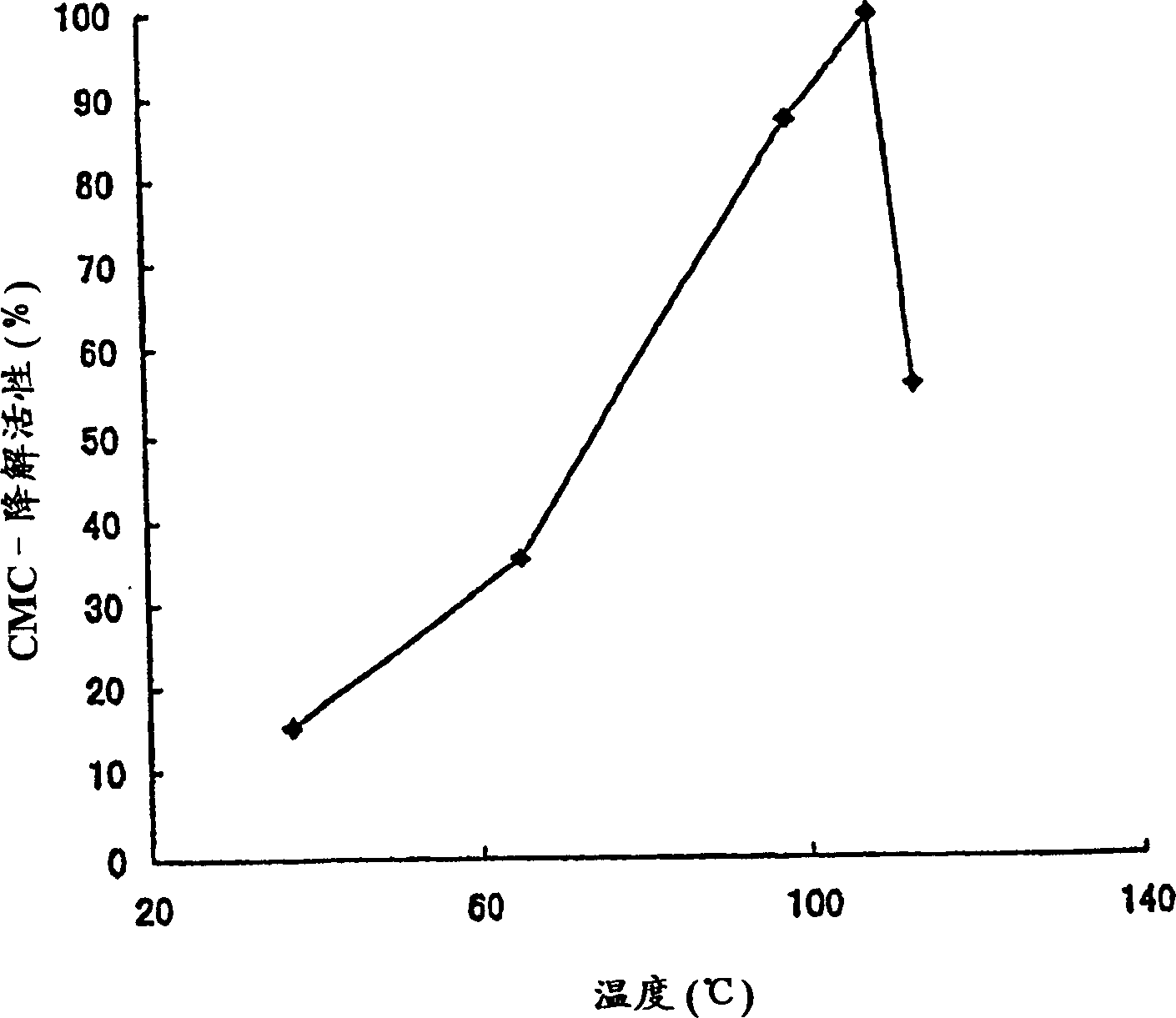
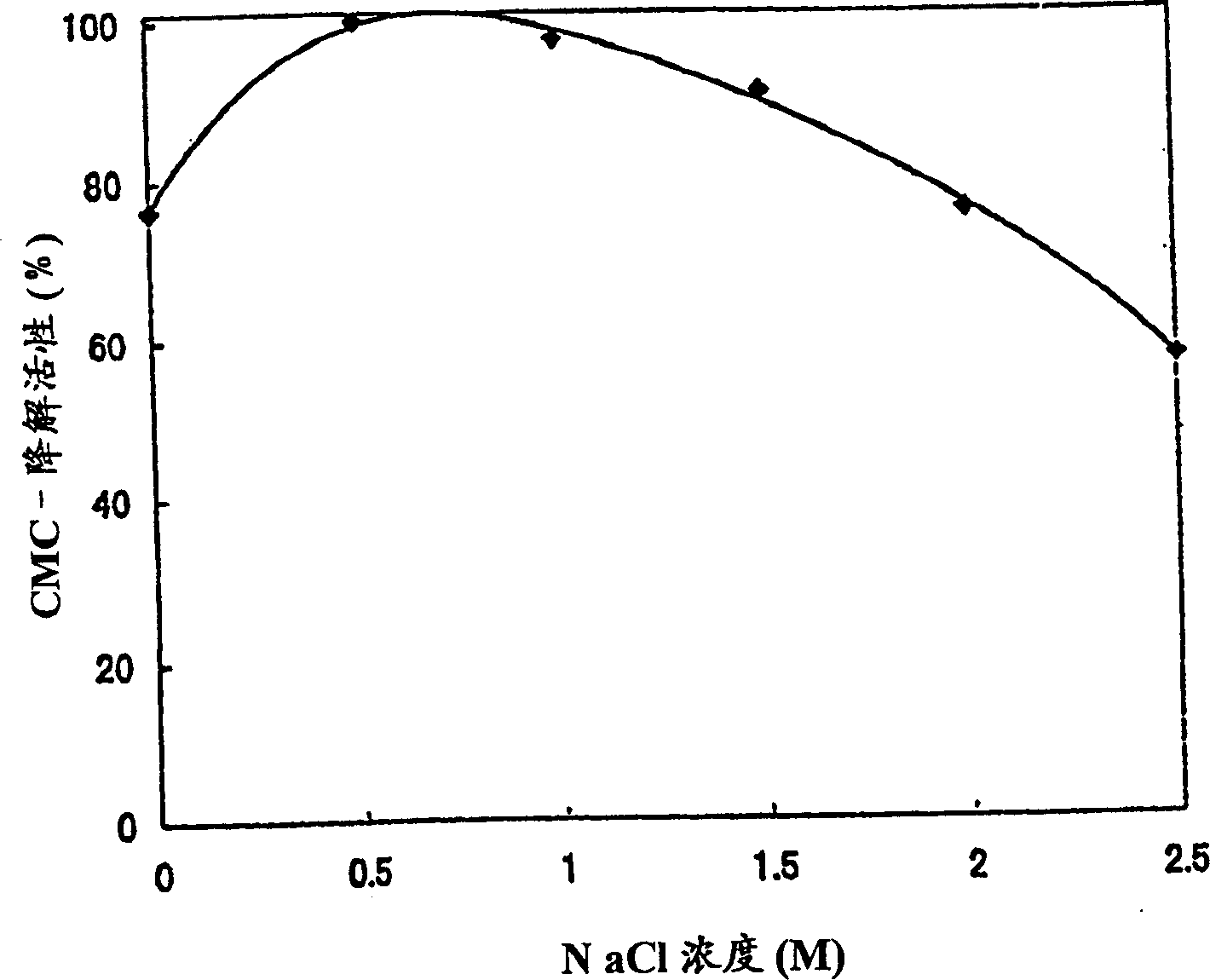
[0123] Physical and Chemical Properties of Cellobiohydrolase Activity of Polypeptides of the Invention
[0124] The polypeptide solution of the present invention prepared in Example 2-(2) was used in this example. Determination of cellobiohydrolase activity with the following activities: 1) according to the Park and Johnson method, by detecting the amount of cellobiose produced using CMC as a substrate as the amount of reducing sugar, the CMC-degrading activity determined; 2) by detecting The amount of cellobiose produced by using pNPC as the substrate is used as the amount of p-nitrophenol, and the p-nitrophenyl-β-D-cellobioside (pNPC)-degradation activity of determination; 3) by detecting the application of 4-MUC 4-Methylumbelliferyl-β-D-cellobioside (pNPC)-degrading activity determined as amount of 4-methylumbelliferyl-β-D-cellobioside (pNPC) as amount of cellobiose produced as substrate .
[0125] (1) Dependence on reaction pH (relationship)
[0126] Prepare buffers as fo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com