Peptide nucleic acid chip for detecting muatatonal site of hepatitis B virus and its preparing method
A hepatitis B virus, mutation site technology, applied in the field of gene chips
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
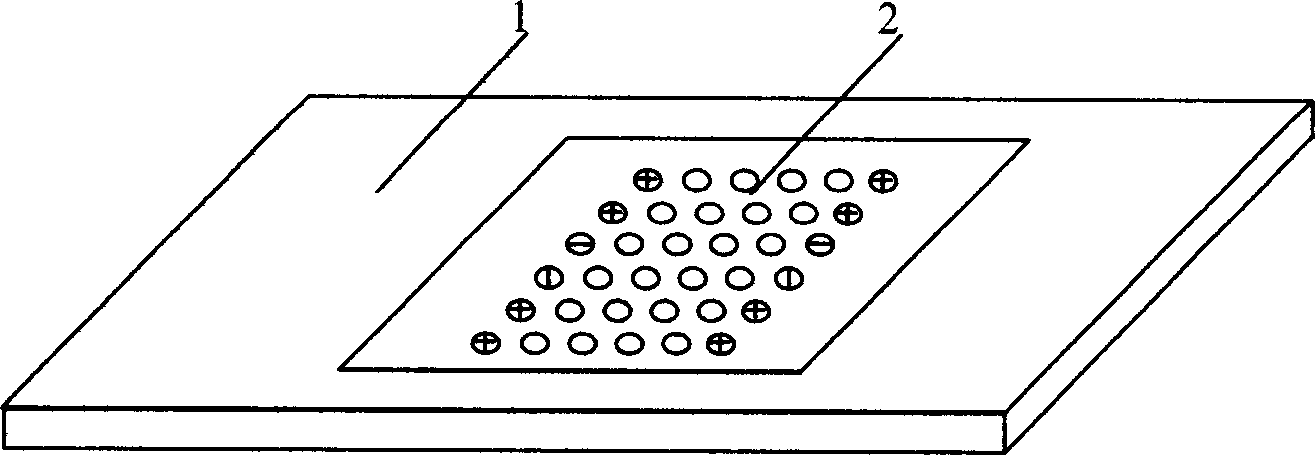
[0047] Embodiment 1: structure as attached figure 1 Shown:
[0048] Peptide nucleic acid probes and control point coating 2 are evenly distributed in an array on the substrate 1, which contains 24 detection probes of known mutation sites and wild sites in the pre-C and S regions of hepatitis B, and 2 are positive Control probe, 1 negative control probe and blank sample solution control. Positive probes are located around the array, which not only serves as a positive control, but also serves as a positive signal to locate the array. The PCR amplified product of the hepatitis B DNA sample is labeled by in vitro transcription and hybridized with the peptide nucleic acid array, and the result is obtained by scanning with a laser confocal scanner, and analyzed and judged.
Embodiment 2
[0049] Example 2: Modification of slides and fixation of peptide nucleic acid probes
[0050] 1. Modification of slides
[0051] The blank glass slide was washed with 10% NaOH, then washed with 1% HCI acid solution, washed with water and then dried to obtain a clean glass bare chip. Treat with 95% acetone solution of 1% 3-aminopropyltrimethoxysilane for 30 minutes, then wash with acetone for 3 times, 5 minutes each time, and bake at 80°C for 15 minutes to obtain the aminated glass slide. Continue to treat with 10% glutaraldehyde aqueous solution for 30 minutes, then wash with double distilled water, and obtain an aldylation-modified glass slide after vacuum drying.
[0052] 2. Immobilization of peptide nucleic acid probes (take the formaldehyde slide as an example)
[0053] The design and selection of peptide nucleic acid probes are shown in Table 1.
[0054] Immobilization of peptide nucleic acid probes on aldylated slides with saturated NaHCO 3 solution as the sample sol...
Embodiment 3
[0061] Example 3: Extraction, PCR amplification and labeling of hepatitis B sample DNA
[0062] 1. Extraction of DNA from hepatitis B samples
[0063] 1) Take 100 μl of hepatitis B serum, add 3 times the volume of lysate (4M guanidine thiocyanate, mercaptoethanol, 0.1M Tris-CI, pH7.5), and mix well.
[0064] 2) Add an equal volume of chloroform and isoamyl alcohol solution (24:1), and mix well. Centrifuge at 12,000 rpm for 15 minutes at room temperature.
[0065] 3) Carefully transfer the supernatant into another 1.5ml centrifuge tube, and add 2 times the volume of absolute ethanol. Store at -20°C for 2 hours. Centrifuge at 12000 rpm for 15 minutes at 4°C.
[0066] 4) The precipitate obtained after centrifugation was washed twice with 70% ethanol, and centrifuged at 12000 rpm for 10 minutes at 4° C. each time.
[0067] 5) The precipitate was naturally dried and dissolved in 16 μl of pure water.
[0068] 2. Amplification and purification of PCR target fragment
[0069] 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com