Method and kit for detecting ox and sheep components in feed
A technology for feed, cattle and sheep, applied in the field of detection, can solve the problems of large quantitative error, cumbersome analysis process, inability to meet the requirements of rapidity and high sensitivity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Design and Synthesis of Primer Pairs and Probes Specific to Bovine / Ovine Components in Feed
[0053] According to the characteristic sequences of various cattle, sheep, pigs, chickens, fish, horses, etc. listed in the NCBI database, after sequence comparison, select a suitable fragment from the D-Loop region, and then use ABI's PrimerExpress software, The following primers and probes with very high specificity were designed for cattle and sheep respectively:
[0054] Upstream primer: 5'GAG CTT AAT TAC CAT GCC G 3' (SEQ ID NO: 3);
[0055] Downstream primer: 5'TTA TGT GTG AGC ATG GGC 3' (SEQ ID NO: 4);
[0056] Bovine probe: FAM-5'TTT CTT CAG GGC CAT C 3'-TAMRA (SEQ ID NO: 5);
[0057] Sheep probe: (FAM-5'AGG GAT CCC TCT TC 3'-TAMRA) (SEQ ID NO: 6);
[0058] Universal probe: (FAM-5'AGG GAT CCC TCT TC 3'-TAMRA) (SEQ ID NO: 7);
[0059] Among them, FAM is a fluorescent reporter group, and TAMRA is a fluorescent quencher group.
[0060] The bovine D-Loop region sequenc...
Embodiment 2
[0063] Detection of ingredients of bovine / sheep origin in feed
[0064] First, extract DNA samples from various feed samples by conventional methods, or use the following extraction kits to extract DNA samples from various feed samples:
[0065] A. Buffer 1 (100mmol / L Tris-HCl, pH=8.0, 0.20mmol / L EDTA, 500mmol / L NaCl)
[0066] B. Buffer 2 (diethyl ether, phenol: chloroform: isoamyl alcohol = 24: 25: 1, 10% SDS)
[0067] C. Wash buffer (70% ethanol, 3mol / L sodium acetate)
[0068] D. Proteinase K (dry powder, 10mg / tube, mixed with 10mg / mL when used)
[0069] E. DNA adsorption column.
[0070] Next, add primers (0.25ul each, concentration 20μM / L) and extracted DNA to a PCR reaction system (40uL) containing the following components:
[0071] A. 20% glycerol (10uL)
[0072] B. 10×TaqMan Buffer A (5uL)
[0073] C. 25uM MgCl 2 (10uL),
[0074] D. dATP(1uL), dCTP(1uL), dGTP(1uL), dUTP(1uL)
[0075] E. Gold Taq polymerase (5U / uL, purchased from ABI company) 0.5uL
[0076] F....
Embodiment 3
[0090] Comparison of experimental results of negative samples and positive samples
[0091] In order to verify the method of the present invention, the same method as in Example 2 was used to detect 20 blind samples respectively. That is, the DNA was first extracted from the finely ground feed samples, and then on the 7000 SequenceDetection System (Applied Biosystems, USA) real-time fluorescence quantitative instrument, the samples were detected with universal probes to determine whether the samples were positive or negative. If the sample is positive, the bovine probe and the sheep probe are further used for detection to determine the composition and content of the sample.
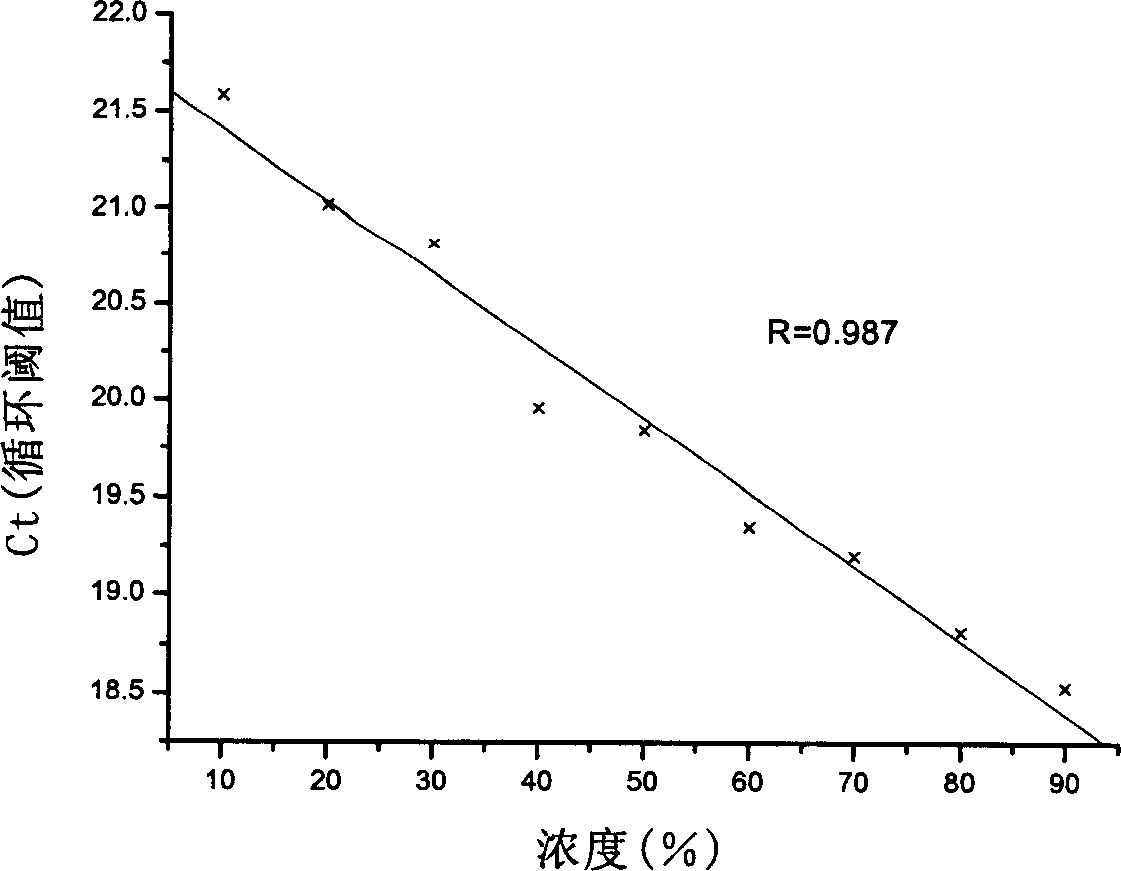
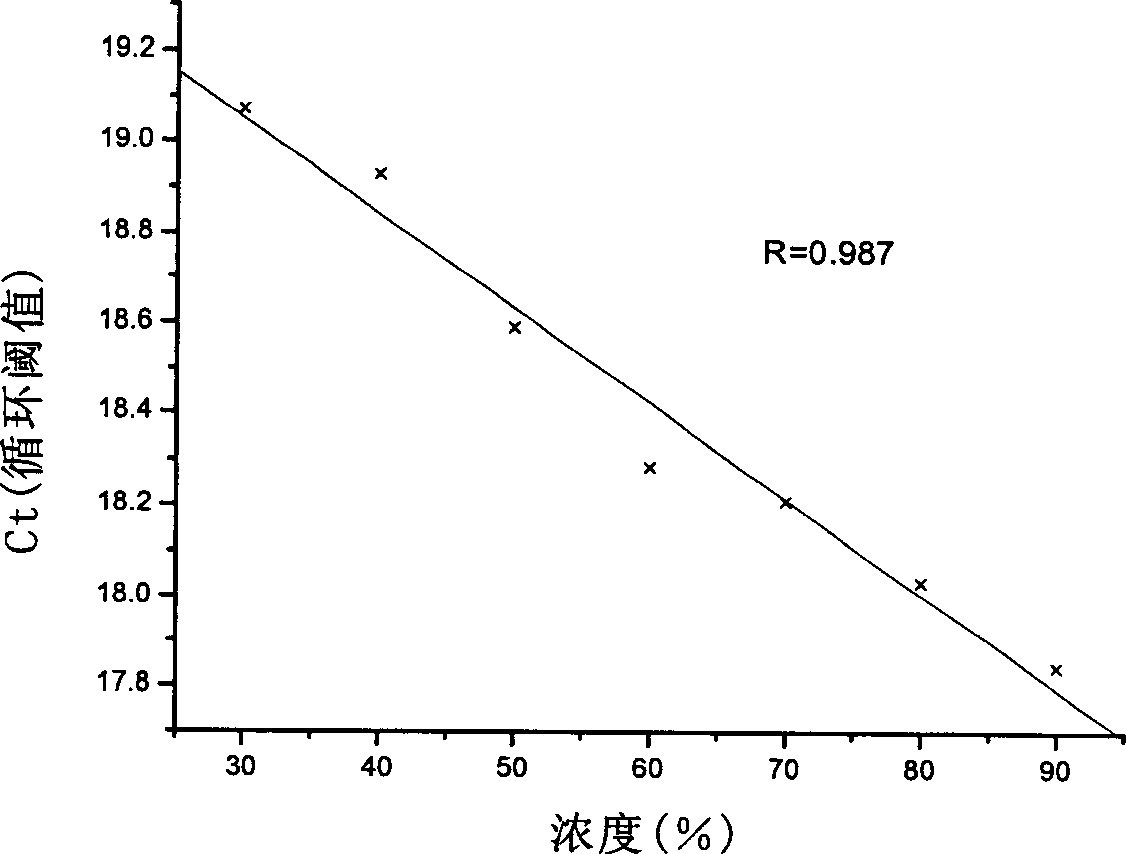
[0092] The test results are shown in Table 3 and Table 4, which are consistent with the actual composition.
[0093]
[0094] Note: % is the mass percentage of feed; Ct value is cycle threshold
[0095]
[0096] Note: % is the mass percentage of feed; Ct value is cycle threshold
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com