Fungus virulence new gene MgKIN17 coming from pyricularia gisea and its use
A virulence, piriformis technology, applied in genetic engineering, plant genetic improvement, application, etc., can solve the problems of unclear differentiation and formation mechanism of appressorium, and uncloned genes.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Screening and identification of mutants.
[0034] a. Preparation of conidia: spread the fully interrupted Pyrospora mycelium evenly on the oatmeal medium plate, and cultivate it at 26°C-28°C. When the new mycelium can be seen growing out of the surface of the medium, Gently wash off the mycelium with a cotton swab, rinse it with water, cover it with a single layer of gauze and culture it under light at 26°C-28°C for about 48 hours.
[0035] b. Preparation of spore suspension: wash the spores described in (1) and filter them in a 50ml centrifuge tube with double-layer lens paper, centrifuge at room temperature for 5 minutes at 4000rpm to collect the spores, and then suspend them in 0.25‰ Tween 20 . Using a hemocytometer and microscope, adjust the concentration of conidia to 10 per mL 4 spore.
[0036] c. Determination of pathogenicity: Spray and inoculate the spore suspension of the wild-type bacterial strain and the mutant evenly on the front of the leaves...
Embodiment 2
[0038] Example 2. Co-segregation analysis of mutant phenotypic changes and insertion markers.
[0039] The above-mentioned mutant H680 and the non-hygromycin-resistant strain S1528 with normal pathogenicity were cultured in confrontation on the tomato oatmeal medium. First cultivate at 25°C until the edges of the colony are about to join together, then move it to 20°C for light culture, and about 20 days form a black protruding ascus at the junction of the colony. Pick the mature ascus shell, squeeze it gently in sterile water, release the ascus in the shell, spread the ascus suspension on the water agar plate, and pick the single hyphae germinated by ascospores in the ascus 24 hours later on the tomato cultured on oatmeal medium. The hygromycin resistance of the progeny of these ascospores was counted, and the germination rate of the spores and the formation rate of appresses, infection nails and infection hyphae were observed and recorded on the onion epidermis. The result...
Embodiment 3
[0042] Example 3 Cloning of genes involved in appressorium formation of Pyricularia spp.
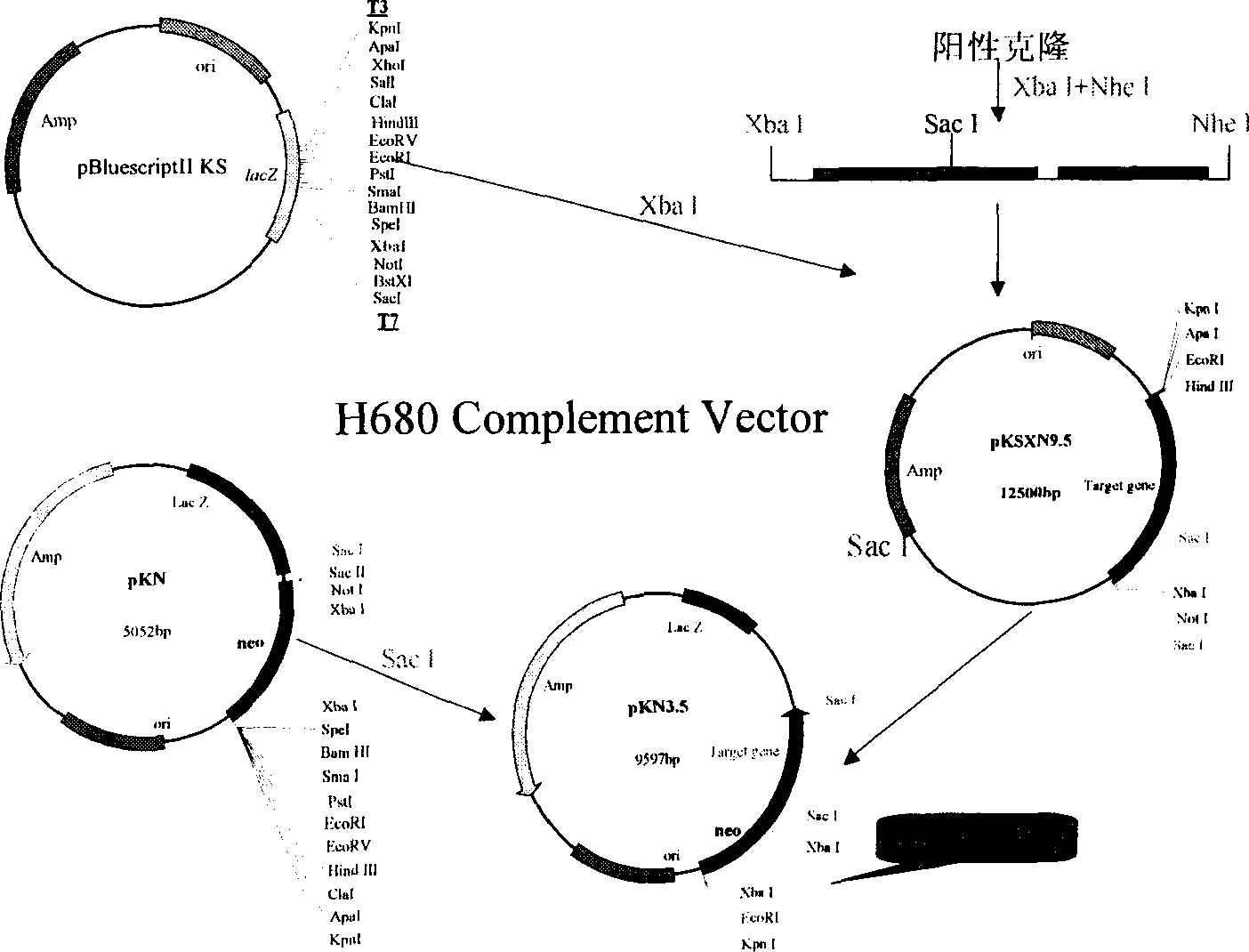
[0043] 1. Plasmid rescue. The genome of the mutant was completely digested with a restriction endonuclease without a restriction site in the inserted plasmid pUCATPH, refined by ethanol precipitation, and then self-ligated, and transformed into competent cells of Escherichia coli. The plasmid of the recombinant transformant was identified by enzyme digestion, in addition to the sequence of pUCATPH, the plasmid also contained part of the genome sequence on both sides. The sequence of these two segments adjacent to the insertion site was compared with the pyrosporium database to determine the disrupted gene ( FIG. 6 ).
[0044] 2. Screening of the genome TAC library: use the partial genome sequence contained in the rescued plasmid as a probe to screen the genome TAC library of Pyrosporium to obtain 8 positive clones, all of which include the full length of the desired gene, and its sequen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com