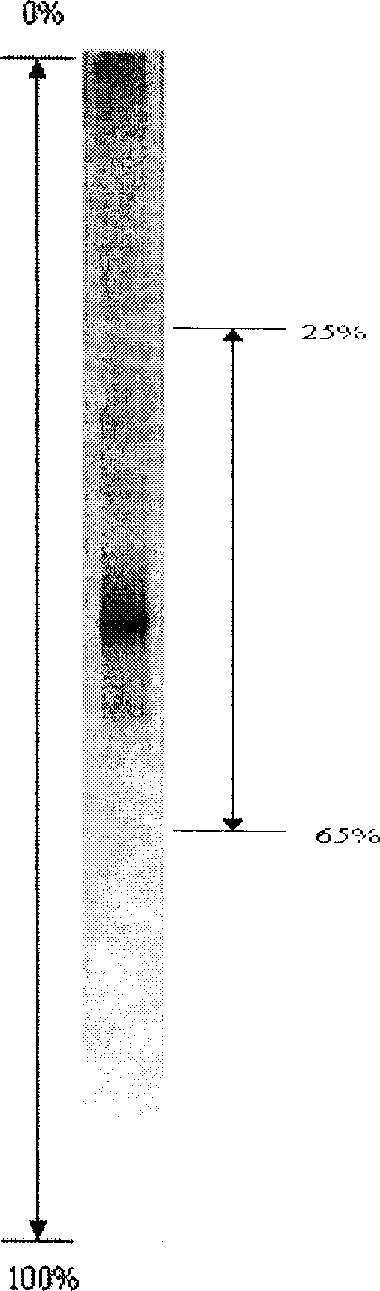
Molecular identification method for structure of dominant bacteria in digestive canal of aquatic animal and its special primer
A technology for dominant flora and aquatic animals, applied in biochemical equipment and methods, measurement/inspection of microorganisms, material separation, etc., can solve problems such as morphological differences, cumbersome operation processes, and the inability of artificial culture of microorganisms, so as to overcome uncertainty Sexuality and accurate results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1. Analysis of the structure of the dominant bacterial flora in the digestive tract of seawater fish-Sichuan snapper (Lutjanus sebae)
[0038] 1. Prepare samples
[0039] Healthy Sichuan snapper (weight: 241.9g) was randomly selected from marine culture cages in Lingshui Xincun Harbor, Hainan Province. The water temperature was 30°C and the seawater salinity was 33‰ when the samples were measured. The experimental fish were fed with single-extruded feed ( Containing 8.2% moisture, 45.0% crude protein and 8.5% crude fat), fed 3-4 times a day, and sampled at 15:10 after feeding at 13:00-14:00 on the sampling day. When sampling, use a dipping net to quickly remove the fish from the cage, beat the head with scissors to kill, then put the fish in a sterilized plastic sealed bag, store it in ice, and transfer it to the laboratory within 3 hours, and within 12 hours Process fish samples.
[0040]Fish samples are processed on a sterile operating table. First, steriliz...
Embodiment 2
[0069] Example 2. Analysis of the structure of the dominant bacterial flora in the digestive tract of freshwater fish-Oreochromis niloticus×O.aureus
[0070] The structure analysis of the digestive tract dominant flora of freshwater fish-Oni tilapia (Oreochromisniloticus×O.aureus) is carried out in the same way as in Example 1. The difference in the method is that when the sample is prepared in step 1, the wall of the digestive tract is rinsed The sterile flushing solution used in the process was changed to sterile water; the primer sequence for amplifying the 16s rDNA v6-v8 region fragment in step 3 was V 6 F1 and V 6 R.
[0071] Results The partial structure of the dominant flora in the intestinal wall of the tilapia was as follows: Aeromonas hydrophila, with a relative abundance of 21.3%; Escherichia coli, with a relative abundance of 12.5%; Photobacterium fluorescens, with a relative abundance of 4.0%. The rest of the dominant flora can be obtained by analogy in the same...
Embodiment 3
[0072] Embodiment 3, Crustacean-vannamei white shrimp (Penaeus vannamei) digestive tract dominant flora structure analysis
[0073] The structure analysis of the digestive tract dominant flora of crustacean-Penaeus vannamei (Penaeus vannamei) is basically the same as in Example 1. The difference in the method is that when preparing the sample in step 1, the sterile bacteria used when washing the digestive tract wall The flushing solution is replaced with sterilized and filtered aquaculture water; the primer sequence for amplifying the 16s rDNA v6-v8 region fragment in step 3 is V 6 F1 and V 6 R.
[0074] Results The partial structures of the dominant flora in the gastric wall of Penaeus vannamei were: Lactobacillus sp., with a relative abundance of 10.2%; Closteridium perfringens, with a relative abundance of 3.2%. The rest of the dominant flora can be obtained by analogy in the same way. The flora structure of other parts of the digestive tract can also be obtained by the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com