Multi-copy monomolecular nucleic acid array chip
A molecular nucleic acid and microarray chip technology, which is applied in the field of biomedical single-molecule nucleic acid array chip manufacturing, can solve the problem of increasing the cost of the chip and the complexity of the preparation process, hindering the use of microarray chip technology, and affecting the accuracy and reliability of the chip and other problems, to achieve the effect of short preparation time, low cost and high preparation cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
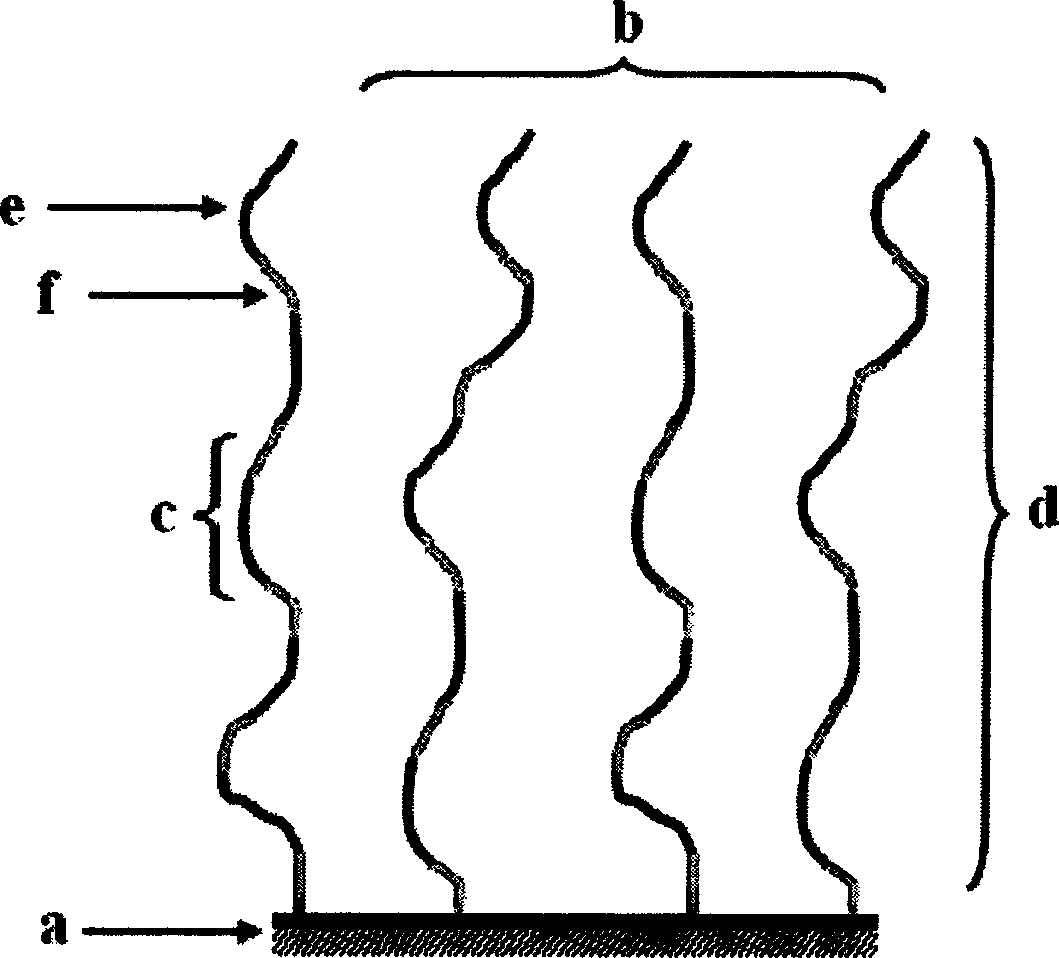
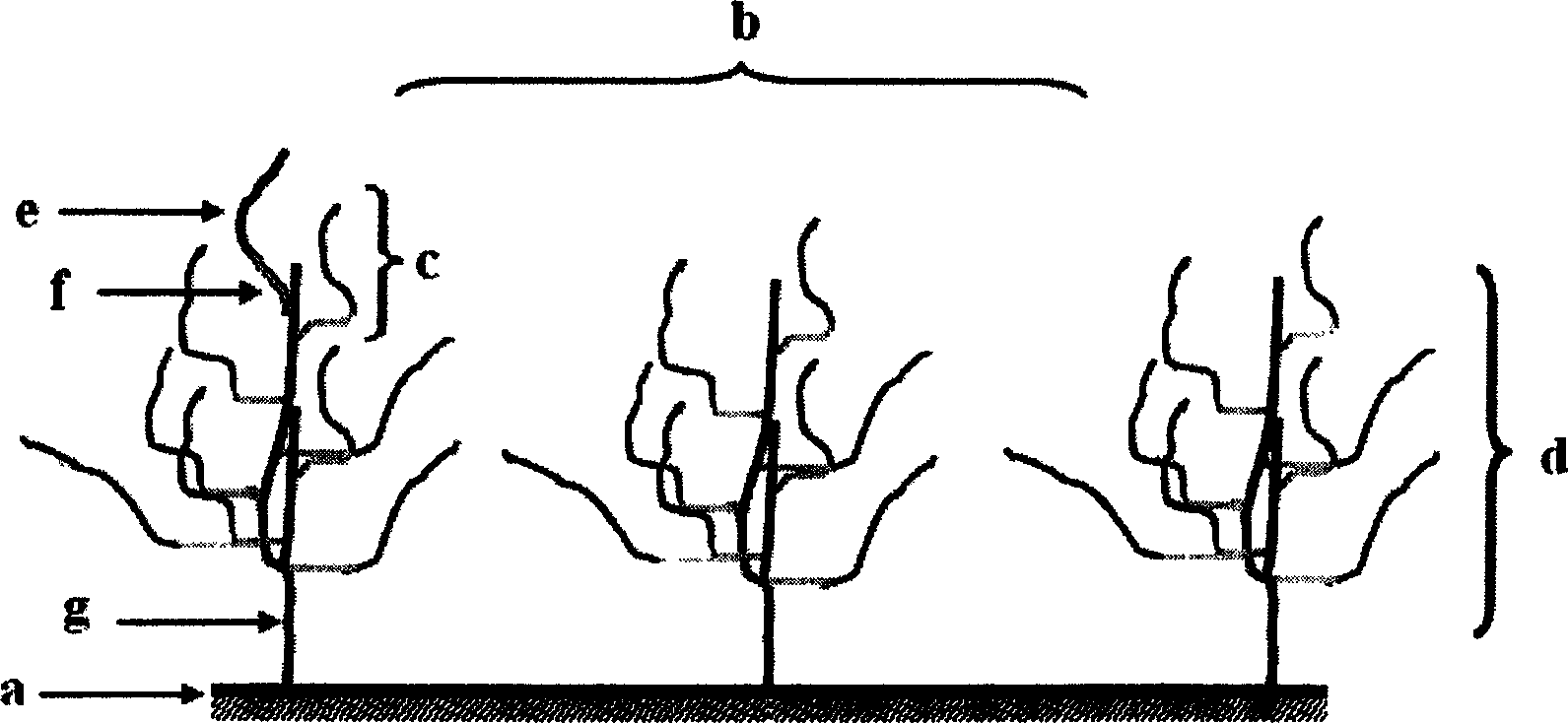
Image
Examples
Embodiment 1
[0033] Example 1. Application of rolling circle amplification technique to prepare single-molecule multi-copy nucleic acid microarray chip.
[0034] ●Selection of enzyme cutting sites and preparation of circular single-stranded genomic DNA fragments
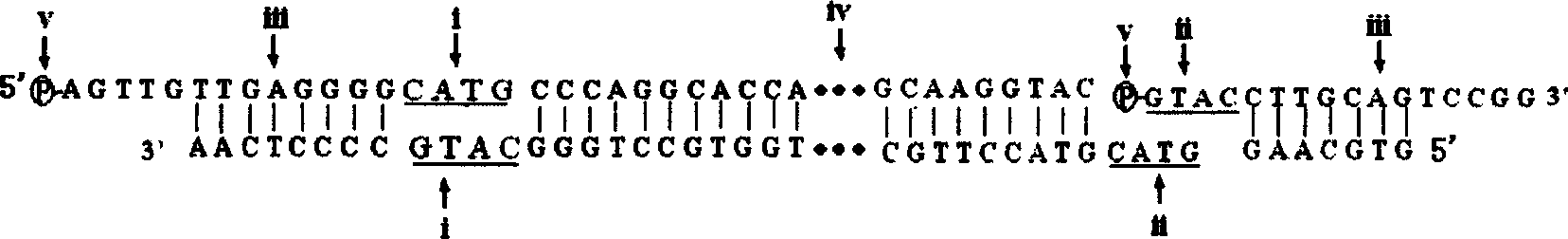
[0035] Restriction enzyme digestion of genomic DNA: firstly, select an appropriate endonuclease (as shown in Table 1) to digest genomic DNA by bioinformatics method, and cut the genome into cohesive end fragments with a size of 50-200 bp. At the same time, according to the characteristics of the end sequence after each endonuclease digestion, design 2 universal linkers. image 3 Taking the combination of Nla III endonuclease and Dpn II endonuclease as an example, the design diagram of the two universal linker sequences. After the human genome DNA extracted from the blood was digested under different restriction endonucleases, 1 μl of the product was electrophoresed on 1.5% agarose gel for 30 minutes to detect the digestion effec...
Embodiment 2
[0043] Example 2, Application of microemulsion technology to prepare single-molecule multi-copy nucleic acid microarray chip
[0044] Microemulsion means that one or several liquids are dispersed in another immiscible liquid in the form of particles (droplets or liquid crystals) to form a multiphase dispersion system with considerable stability. Because their appearance is often milky, so called an emulsion. Its main features are thermodynamic stability, isotropy, transparent or translucent appearance, uniform particle size from nanometer to micron level. Microemulsion related technology is very common in chemistry. We use its thermodynamic stability and other characteristics to place biochemical reactions such as PCR amplification in microemulsion droplets, so that the reaction can be carried out efficiently and quickly.
Embodiment approach
[0045] Concrete implementation scheme is as follows:
[0046] ●Single-molecule parallel PCR in microemulsions
[0047] The preparation of water-in-oil microemulsion at first selects suitable oil phase (mineral oil etc.) and surfactant (Tween, SDS, Triton etc.), is water phase with parallel PCR amplification system, oil phase and water phase by 20: Mix at a ratio of 1 to 1:1, and prepare microemulsions by shaking or ultrasonication. The particle size and uniformity of the prepared microemulsion were observed under a microscope. By controlling the ratio of the water phase and the oil phase, the preparation method, the preparation temperature, the concentration of the surfactant and other conditions, a microemulsion system with uniform particle size can be obtained, and the particle size of the prepared microemulsion can be further optimized to be suitable for polymer molecular microemulsion. Basic single-molecule multi-copy amplification system.
[0048] Parallel PCR amplific...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com