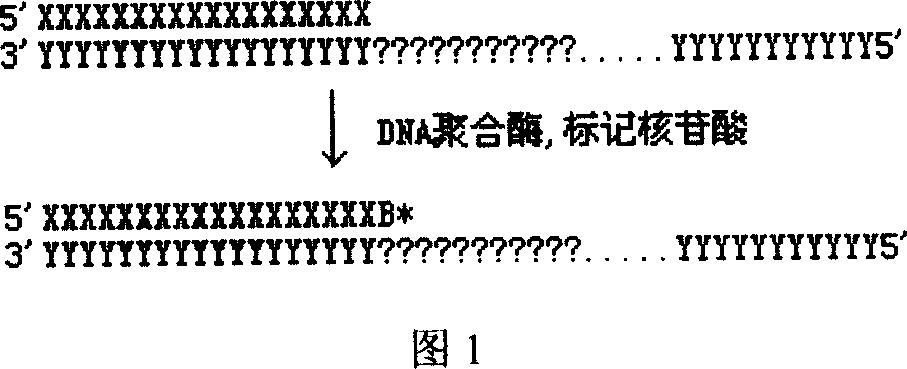
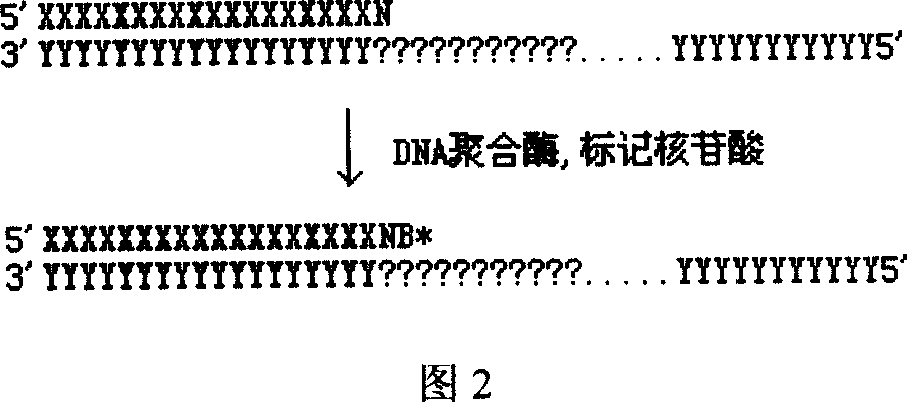
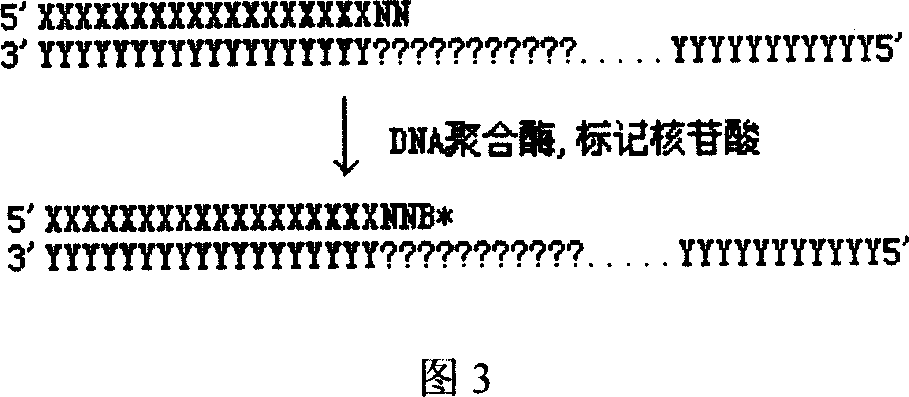
DNA sequence measurement based on primer extension
A DNA sequence and determination method technology, applied in the field of DNA sequence determination, can solve problems such as background interference, interference with fluorescence intensity measurement accuracy, complex cleavage groups, etc., and achieve the effect of high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] A nucleic acid sequence determination method for determining an unknown nucleic acid sequence:
[0027] A) Introduce a known universal DNA sequence into the DNA template to be tested, and fix it on a solid-phase substrate; B) Design 1-15 oligonucleotide DNA primer combinations, so that it contains a section and the above-mentioned addition The sequence that is complementary to the common DNA sequence, and make the 3' ends of the 1-15 oligonucleotide DNA primers contain 0, 1, 2, ..., 14 degenerate nucleotides respectively; C) according to the The position of the nucleotide site, select a primer from the above primer combination, and hybridize it with the fixed DNA template described in A, so that the nucleotide site to be tested is next to the 3' end of the primer; D) use DNA polymerase and dideoxynucleotide substrates labeled with four distinguishable markers or four deoxynucleotides or dideoxynucleotides labeled with one or more markers respectively, hybridized to the ...
Embodiment 2
[0045] Example 2: Sequencing the artificially synthesized DNA template
[0046] The following DNA sequences were synthesized by Invitrogen:
[0047] Seq1: 5'NH2-(T)25 TT CT C AGG TAG AGT TGG AGC TCA GGA CAT GGC AT
[0048] Seq2: 5'NH2-(T)25 TT AC G TGC GTC AGT TGG AGC TCA GGA CTG GGC TG
[0049] Seq3: 5'NH2-(T)25 TT GT C AGA CGA AGT TGG AGC TCA GGA ATA CTT AT
[0050] Seq4: 5'NH2-(T)25T T AGT CCT AGT TGG AGC TCA GGA TCC TTG GA
[0051] Seq5: 5'-CTGAGCTCCAACT 3'
[0052] Seq6: 5'-TGAGCTCCAACTN 3'
[0053] Seq7: 5'-GAGCTCCAACTNN 3'
[0054] Seq8: 5'-GAGCTCCAACTNNN3'
[0055] Seq9: 5'-AGCTCCAACTNNNN3'
[0056] Seq10: 5'-GCTCCAACTNNNNN 3'
[0057] Sequ11: 5'-CTCCAACTNNNNNN 3'
[0058]Seq1-seq4 are used as DNA templates, and seq5-seq11 are sequencing primers. The oblique and bold part of the template sequence is the primer binding region, the underlined part is the sequence to be tested, and NH2 represents amino modification. N is a degenerate nucleotide, and ...
Embodiment 3
[0079] Example 3: Sequencing of T7 phage DNA library clones
[0080] Materials: T7 bacteriophage DNA was purchased from Shanghai Bioengineering Company; Tsp509 I was purchased from NEB Company; pUC118 vector, competent bacteria and transformation kit, T4 DNA ligase were purchased from Dalian Bao Biological Company; klenow DNA polymerase was purchased from Fermentas Company; 序列订购自invitrogen公司:M13前引物:NH-CAGGAAACAGCTATGAC(seq12);M13后引物:GTAAAACGACGGCCAGT(seq13);测序引物:GCATGCCTGCAGGTCAATT(seq14);CATGCCTGCAGGTCAATTN(seq15);ATGCCTGCAGGTCAATTNN(seq16);ATGCCTGCAGGTCAATTNNN(seq17);TGCCTGCAGGTCAATTNNNN (seq18);
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com