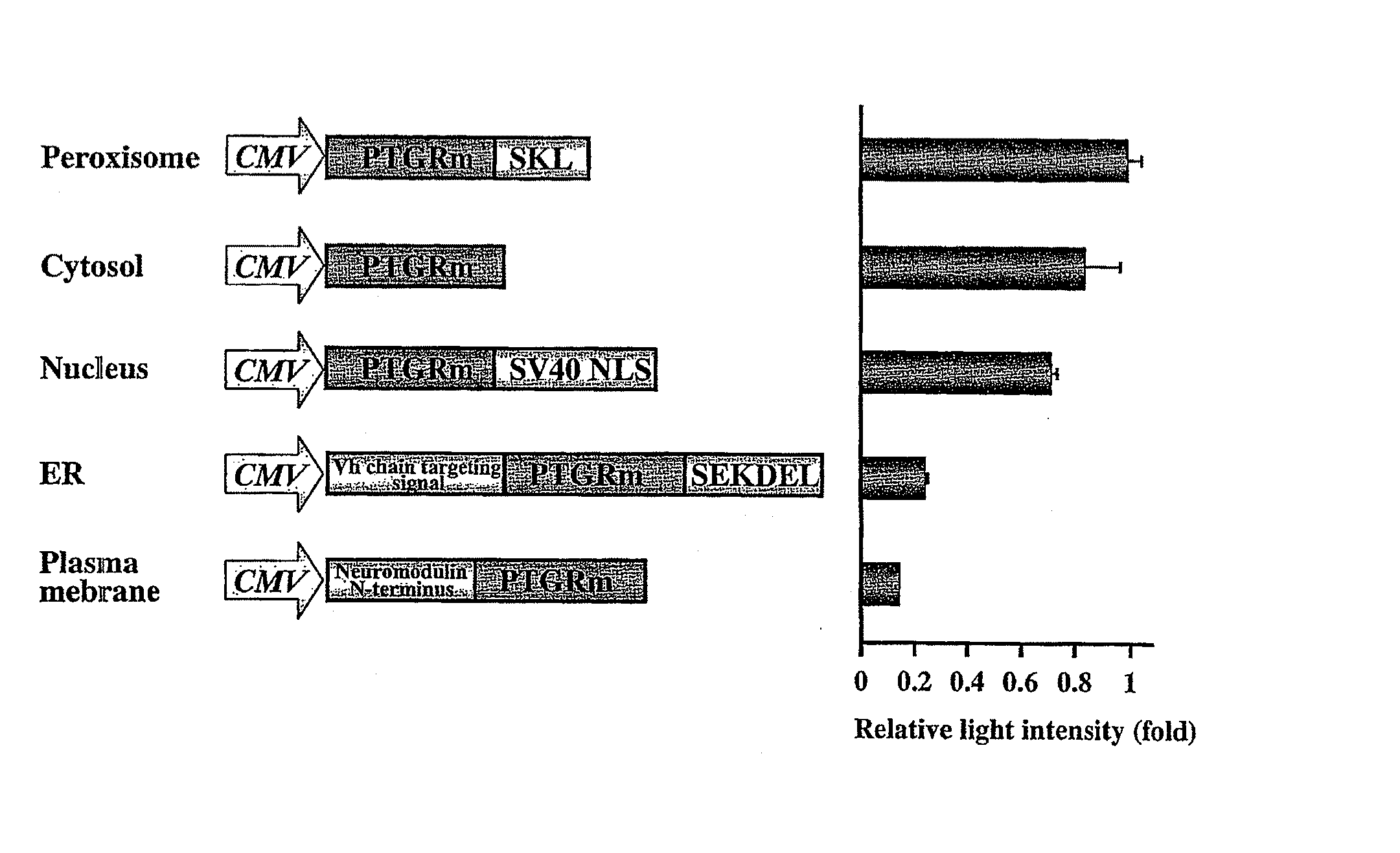
Luciferase gene optimized for use in imaging of intracellular luminescence
a luciferase and luminescence imaging technology, applied in the field of luciferase gene constructs, can solve the problems of inability to observe fluorescent proteins long-term, difficult quantification, impaired cells, etc., and achieve the effects of improving sensitivity, low gene introduction efficiency, and wide enhancement of intracellular amount of expressed luciferas
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
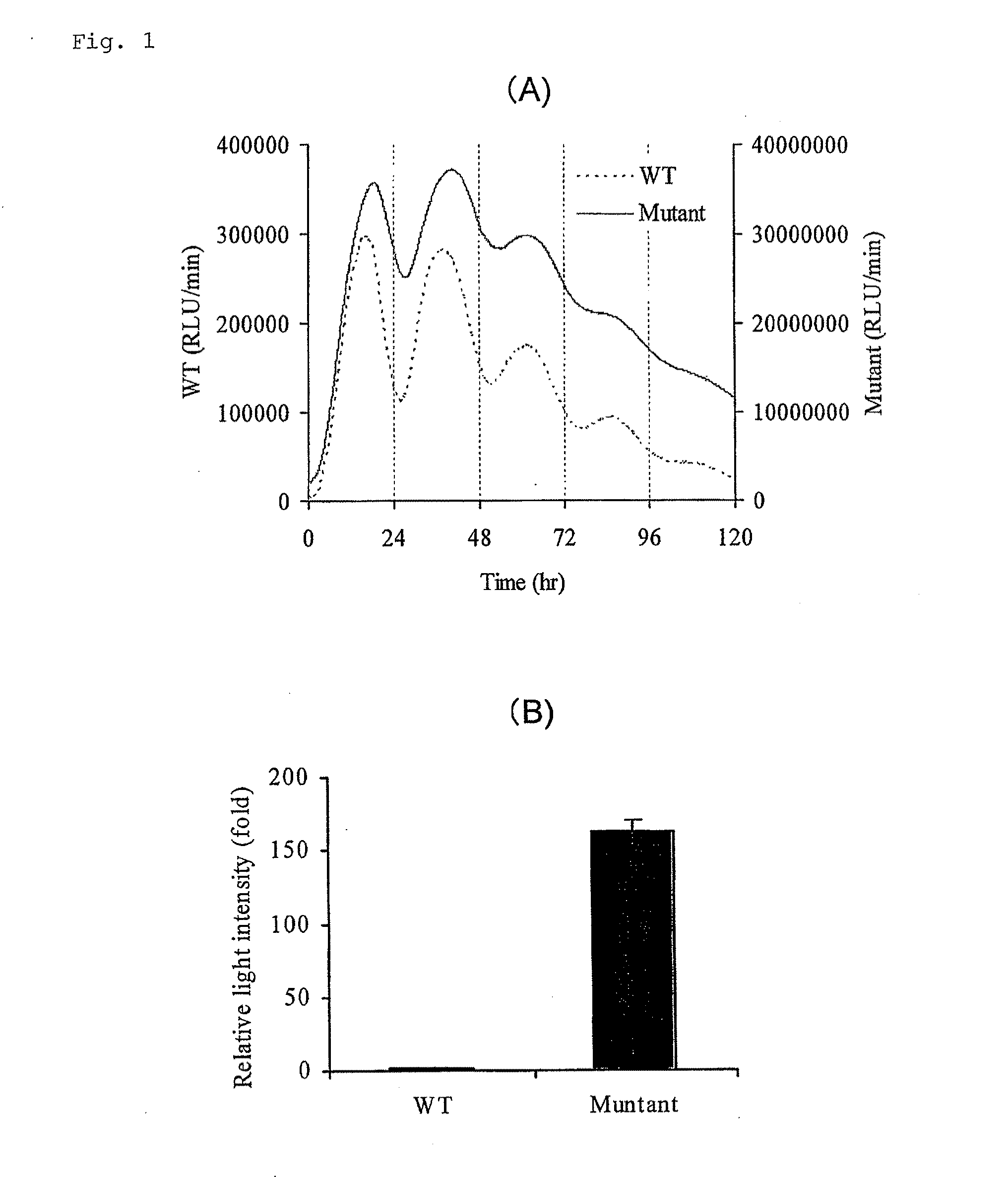
[0108] Short lifespan-type luciferase was made by ligating a PEST sequence (SEQ ID NO: 7) of murine ornithine decarboxylase to a wild-type (SEQ ID NO: 1) and improved-type (SEQ ID NO: 2) click beetle luciferase cDNA. Vectors in which these had been inserted downstream of a murine circadian clock gene Bmal1 promoter (GenBank Accession No. AB064982) were made. Subsequently, 1 μg of each vector was introduced into cultured fibroblast NIH3T3 cells seeded in a 35 mm culture dish by a lipofection method (LipofectAMINE PLUS, Invitrogen), and the cells were cultured at 37° C. for 24 hours and treated with a DMEM medium containing 100 nM dexamethasone for 2 hours. The medium was replaced with a DMEM medium containing 200 μM D-luciferin and 10% (w / v) bovine serum, and then one minute of luminescence was measured every 15 minutes for 5 days in real time using a real-time gene expression measurement apparatus (AB2500 supplied from ATTO Corporation) (FIG. 1A). The change in about a 24-hour cycle...
example 2
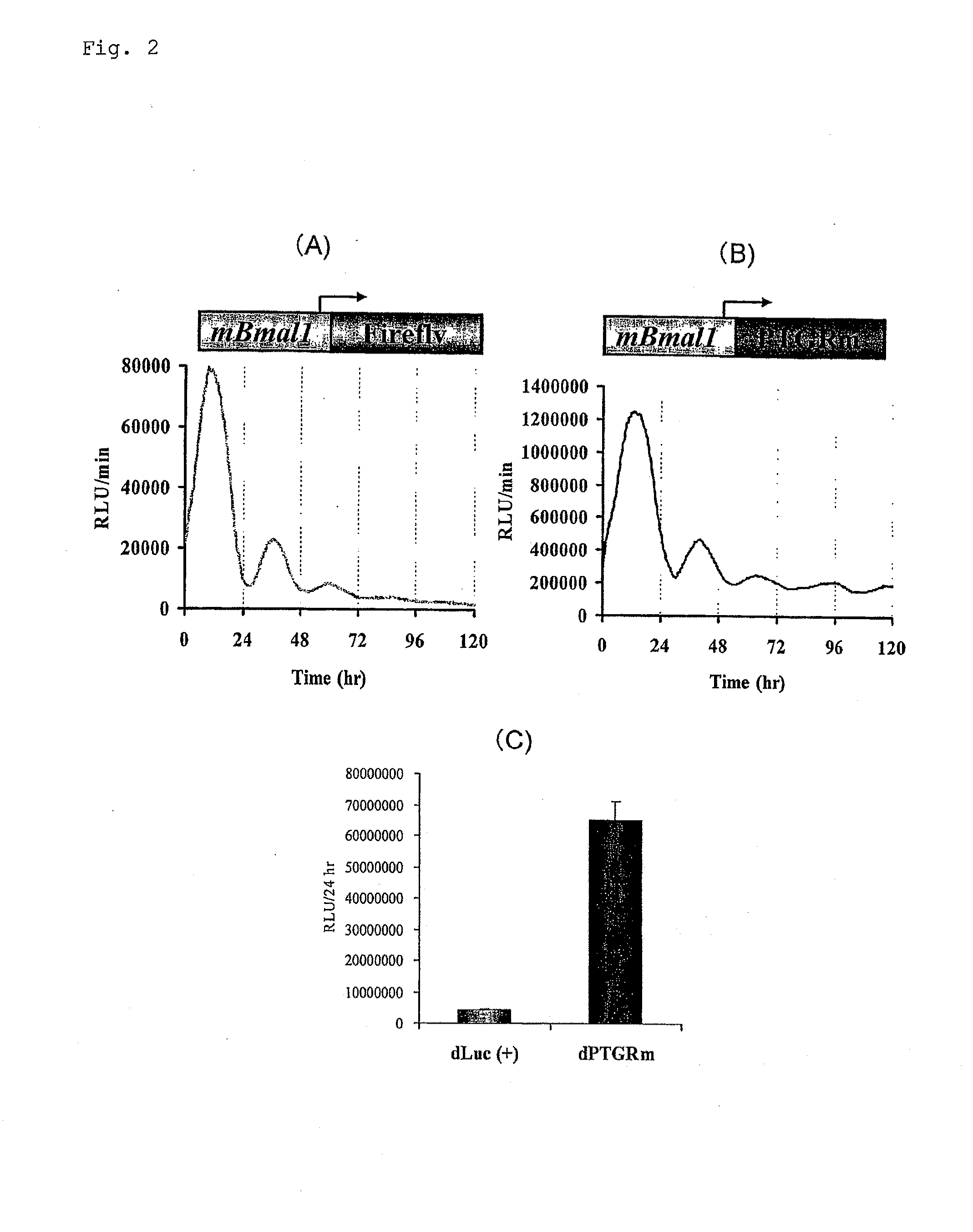
[0110] Short lifespan-type luciferase was made by ligating the PEST sequence (SEQ ID NO: 7) of murine ornithine decarboxylase to firefly luciferase cDNA (Luc(+), supplied from Promega) derived from Photinus pyralis produced in North America and improved-type click beetle luciferase cDNA (SEQ ID NO: 2). Vectors in which these had been inserted downstream of a murine circadian clock gene Bmal1 promoter (GenBank Accession No. AB064982) were made. Subsequently, 1 μg of each vector was introduced into cultured fibroblast rat1 cells seeded in a 35 mm culture dish by the lipofection method (LipofectAMINE PLUS), and the cells were cultured at 37° C. for 24 hours and treated with the DMEM medium containing 100 nM dexamethasone for 2 hours. The medium was replaced with a DMEM medium containing 200 μM D-luciferin and 10% (w / v) bovine serum, and then one minute of luminescence was measured every 15 minutes for 5 days using the real-time gene expression measurement apparatus (AB2500 supplied fro...
example 3
[0111] Expression vectors were made by inserting the aforementioned short lifespan-type firefly luciferase cDNA and short lifespan-type improved-type click beetle luciferase cDNA downstream of an SV40 promoter. The expression vector (200 ng) was introduced into the cultured fibroblast NIH3T3 cells seeded in a 24-well plate by the lipofection method, and the cells were cultured at 37° C. for 48 hours. Subsequently, the medium was replaced with the medium containing 100 μM protein synthesis inhibitor, cycloheximide, and after culturing for 30 minutes, the intracellular luminescence intensity was measured every one hour. The luminescence intensity was measured in the same way as in Example 1 (FIG. 3). In short lifespan-type firefly luciferase derived from Photinus pyralis produced in North America, the half life was one hour; however, in short lifespan-type improved click beetle luciferase, the half life was 4 hours, a fourfold extension.
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| luminescence wavelength | aaaaa | aaaaa |
| luminescence wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com