Gamma-butyrobetaine hydroxylase originated from neurospora crassa
一种碱羟化酶、丁基的技术,应用在γ-丁基甜菜碱羟化酶领域,能够解决副作用等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0038] Example 1: Construction of Neurospora crassa cDNA library.
[0039] To obtain the cDNA of N. crassa, mRNA was first isolated from N. crassa, and cDNA was synthesized from the isolated mRNA by PCR (polymerase chain reaction) using polythymidine (polyT) primers. The cDNA was inserted into the EcoR1 / XhoI site of the AD5 cloning vector, and a cDNA library was constructed in the plasmid as follows. Escherichia coli BNN322 strain was cultured overnight in LB medium supplemented with kanamycin and 0.2% maltose, collected by centrifugation, and dissolved in 1ml of 10mM MgSO 4 in suspension. The bacterial suspension was mixed with 3.5×10 of the constructed cDNA library 7 The lambda phages were incubated together for 30 minutes at 30°C without stirring. Add 2 ml of LB medium to the culture, and incubate the infected strain at 30 °C for 60 min while stirring. The final culture was plated on LB plates containing ampicillin (75 μl- / ml). Plasmids were isolated from the grown col...
example 2
[0040] Example 2: Preparation of primers for obtaining γ-butylbetaine hydroxylase gene.
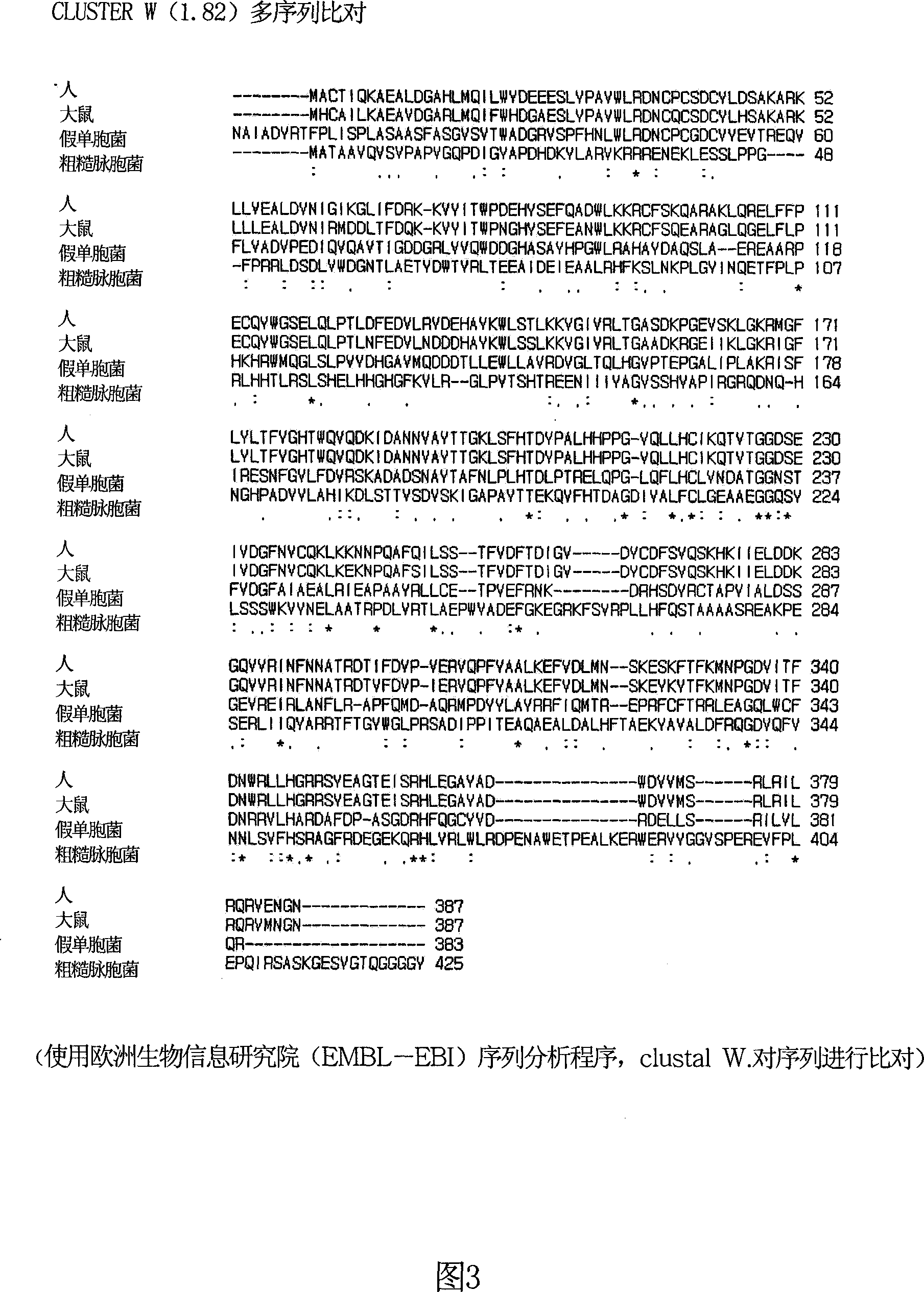
[0041] The amino acid sequence of γ-butylbetaine hydroxylase (γ-BBH) produced by N. crassa was compared with that of γ-BBH produced by human, rat and Pseudomonas ( FIG. 3 ). SEQ ID NO: 1 represents the amino acid sequence of γ-BBH produced by Neurospora crassa, SEQ ID NO: 2 represents the amino acid sequence of γ-BBH from human, SEQ ID NO: 3 represents the amino acid sequence of γ-BBH from murine, and SEQ ID NO: 4 represents the γ-BBH from Pseudomonas - the amino acid sequence of BBH. The sequence homology results are as follows (starting with a pairwise alignment):
[0042] Sequence (1:2) permutation, score: 11%;
[0043] Sequence (1:3) alignment, score: 11%;
[0044] Sequence (1:4) alignment, score: 10%;
[0045] Sequence (2:3) alignment, score: 88%;
[0046] Sequence (2:4) alignment, score: 29%;
[0047] Sequence (3:4) alignment, score: 29%.
[0048] The γ-BBH produced by Neuros...
example 3
[0054] Example 3: Obtaining the gene encoding γ-BBH.
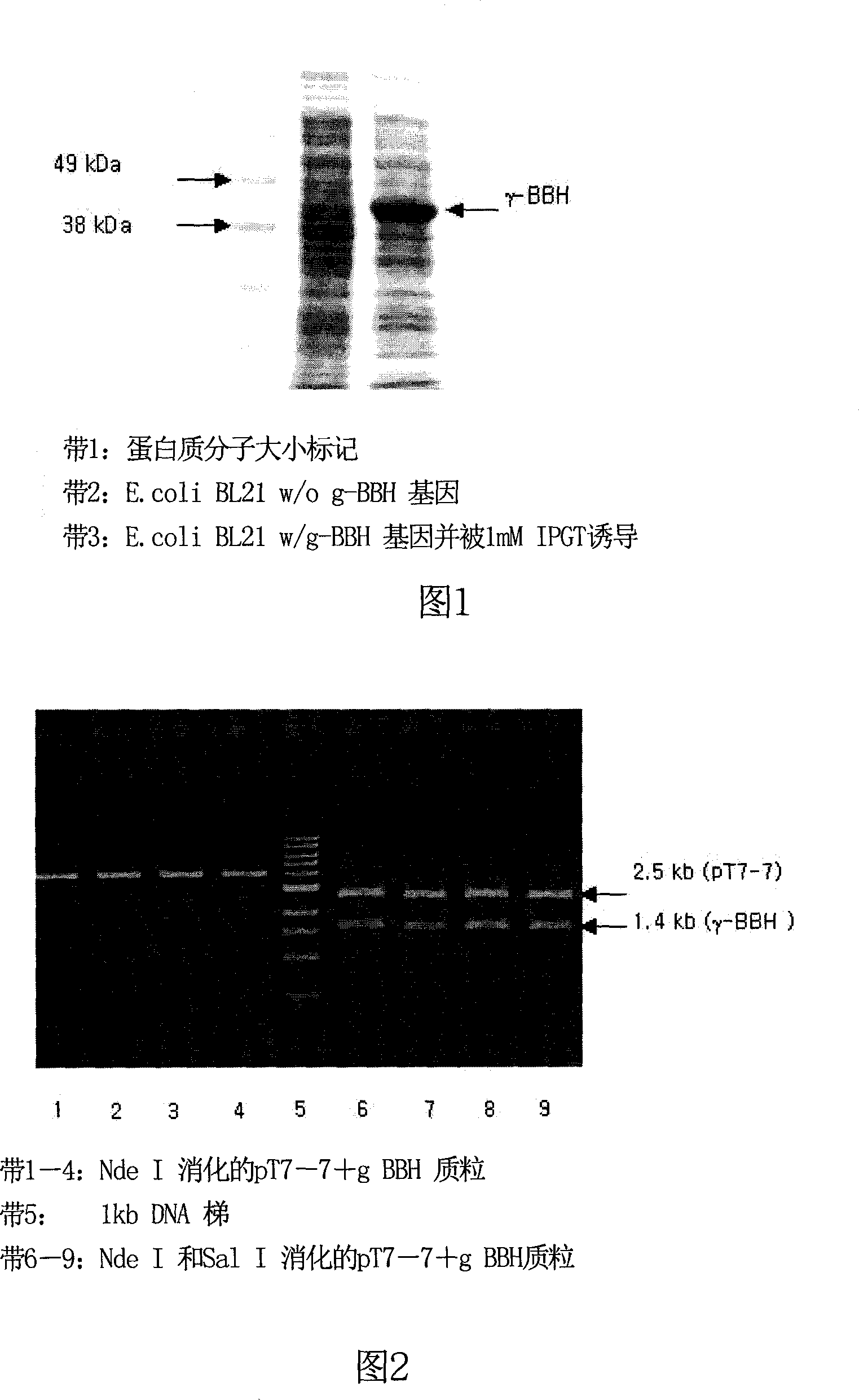
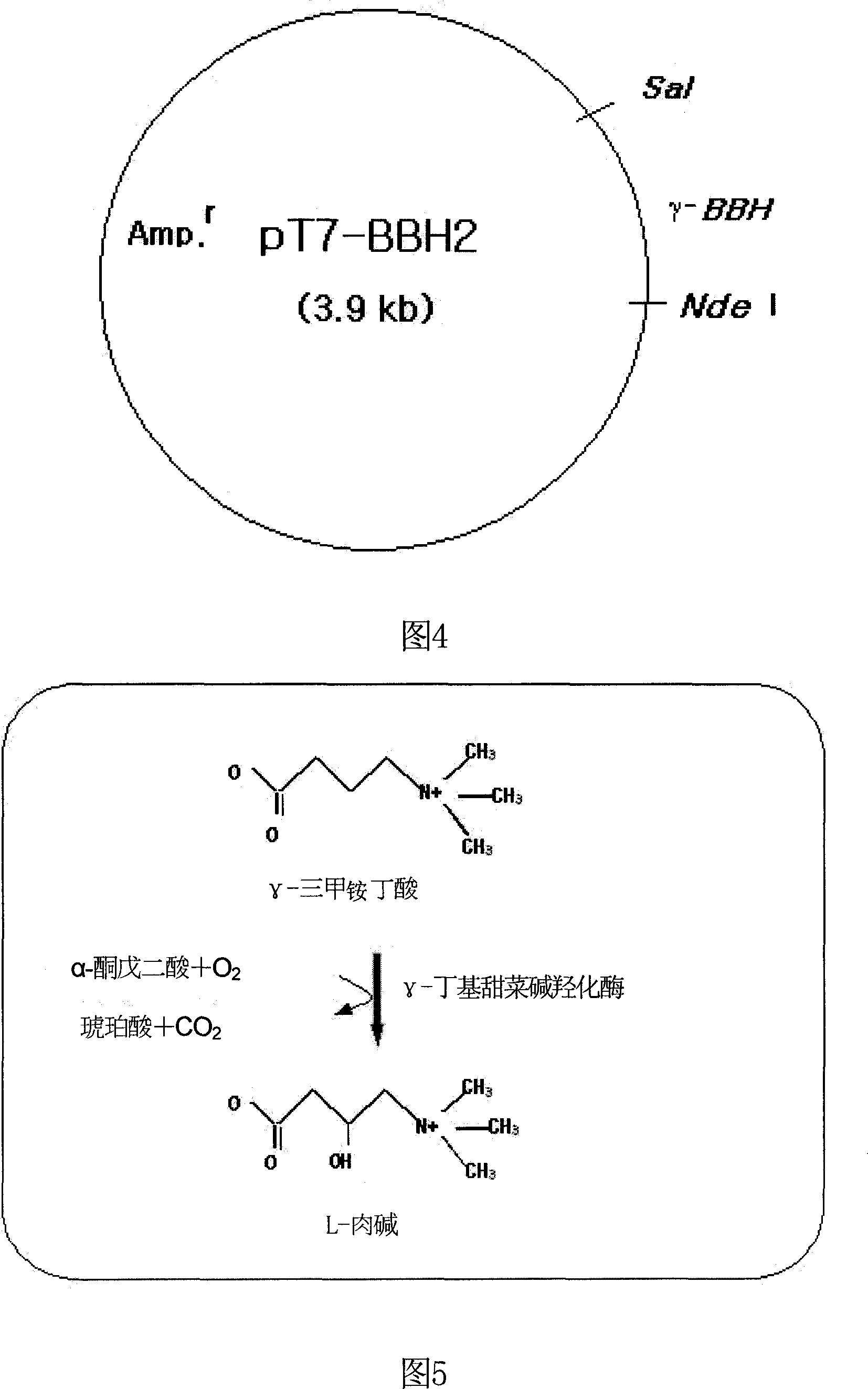
[0055] From the Neurospora crassa cDNA library prepared in Example 1, the γ-BBH gene was amplified by the PCR method using a pair of primers prepared in Example 2. The PCR product was electrophoresed on an agarose gel, and a band was observed at about 1.4 kb. The nucleotide sequence of the amplified gene was determined using an automatic DNA sequencer. At the same time, the BLAST program of NCBI Company was used to perform homology search on the determined nucleotide sequences. As a result, a gene 100% identical to the amplified gene was found in the genome sequence of Neurospora crassa, and the function of the translation product of the found gene was only a hypothetical protein. The PCR product was then digested with EcoRI and Sal I, ligated with pUC19 digested with the same restriction enzymes, and introduced into E. coli DH5. Transformants were identified using blue / white screening. When the plasmid was isolated fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com