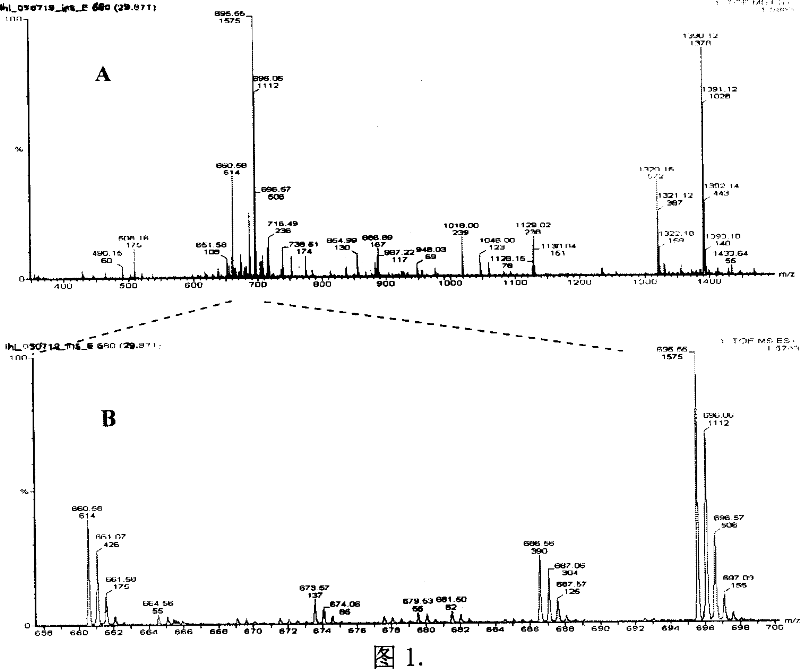
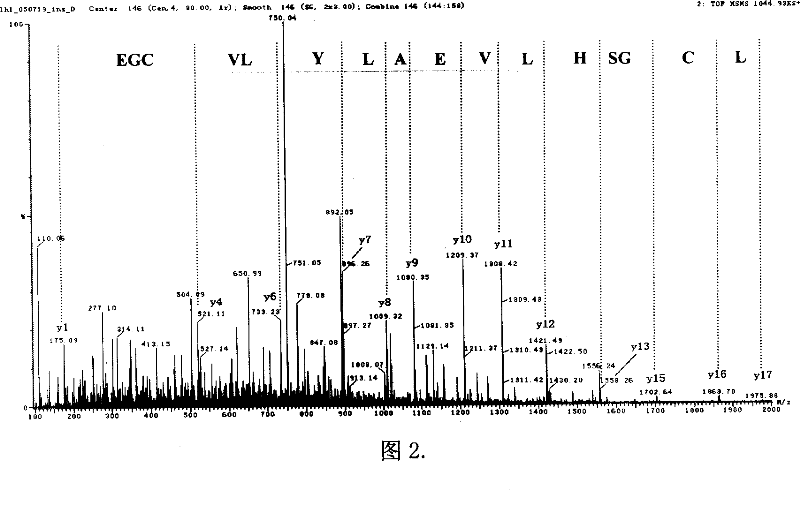
Method and reagent kit for rare-earth element metal chelating making quantitative proteome
A rare earth element and metal chelation technology, applied in the direction of measuring devices, instruments, scientific instruments, etc., to achieve the effect of convenient use, wide practicability, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] The relative quantification of embodiment 1 standard protein insulin
[0033] 1.1 Take 0.1 mg of insulin and dissolve it in 100 μL of 50 mM pH 7.0 triethylamine bicarbonate buffer solution. Add 0.2 μL of 100 mM reducing agent tricarboxyethyl phosphine (TCEP), react at 37°C for 4 hours, then cool to room temperature, add 0.5 μL of 100 mM iodoacetamide (IAA), react at room temperature for 1 hour, and then add 10 mM DTT to terminate the reaction. Add 2ug trypsin, digest at 37°C for 2 hours. Then add 2ug of trypsin, and digest at 37°C for 10 hours. The insulin peptide mixture obtained by enzymatic digestion was added to 360 μg of diethylenetriaminepentaacetic acid dianhydride solid powder, vigorously vortexed for 1 min, and then left at room temperature for 1 hour. Then put the reaction solution in a vacuum dryer and dry it completely, add 0.1M ammonium acetate solution to redissolve, and divide it into two parts according to the volume, and add 40 μL of YCl with a concen...
Embodiment 2
[0036] Embodiment 2 Relative Quantification of Standard Protein Horse Myoglobin
[0037] 2.1 Take 0.1 mg of horse myoglobin and dissolve it in 100 μL of 50 mM pH 7.0 triethylamine bicarbonate buffer solution. Myoglobin has no cysteine residues in its amino acid sequence, so a reductive alkylation step is not required. Add 2ug trypsin, digest at 37°C for 2 hours. Then add 2ug of trypsin, and digest at 37°C for 10 hours. The myoglobin peptide mixture obtained by enzymatic digestion was added to 360 μg of diethylenetriaminepentaacetic acid dianhydride solid powder, vigorously vortexed for 1 min, and then left at room temperature for 1 hour. Then put the reaction solution in a vacuum dryer and dry it completely, add 0.1M ammonium acetate solution to redissolve, and divide it into two parts on average, one of which is added with 40 μL of YCl with a concentration of 0.15M 3 , add the same concentration and volume of TbCl to the other 3 aqueous solution. Incubate at 37°C for 2...
Embodiment 3
[0040] The relative quantification of embodiment 3 six kinds of standard protein mixtures
[0041] 3.1 Configure six protein solutions (bovine serum albumin, transferrin, α-lactoglobulin, β-lactoglobulin, myoglobin and lysozyme protein) with a concentration of 0.1mmoL / L for each protein, The buffer solution was 50 mM triethylamine bicarbonate pH 7.0. Take 70 μL of each and mix together. Add 0.8 μL of 500 mM reducing agent tricarboxyethylphosphine (TCEP), react at 37°C for 4 hours, then cool to room temperature, add 1 μL of 1M iodoacetamide (IAA), and react at room temperature for 1 hour in the dark. Add 5ug of trypsin, digest at 37°C for 2 hours. Then add 5ug of trypsin, and digest at 37°C for 10 hours. The myoglobin peptide mixture obtained by enzymatic digestion was added to 360 μg of diethylenetriaminepentaacetic acid dianhydride solid powder, vigorously vortexed for 1 min, and then left at room temperature for 1 hour. Then put the reaction solution into a vacuum dryer ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com