Recombinant carrier containing endo-chitinase genes
A chitinase and carrier technology, applied in the field of genetic engineering, can solve the problems of high energy consumption, difficult control of the degree of degradation, and secondary environmental pollution for acid degradation, and achieve the effect of industrialized production.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
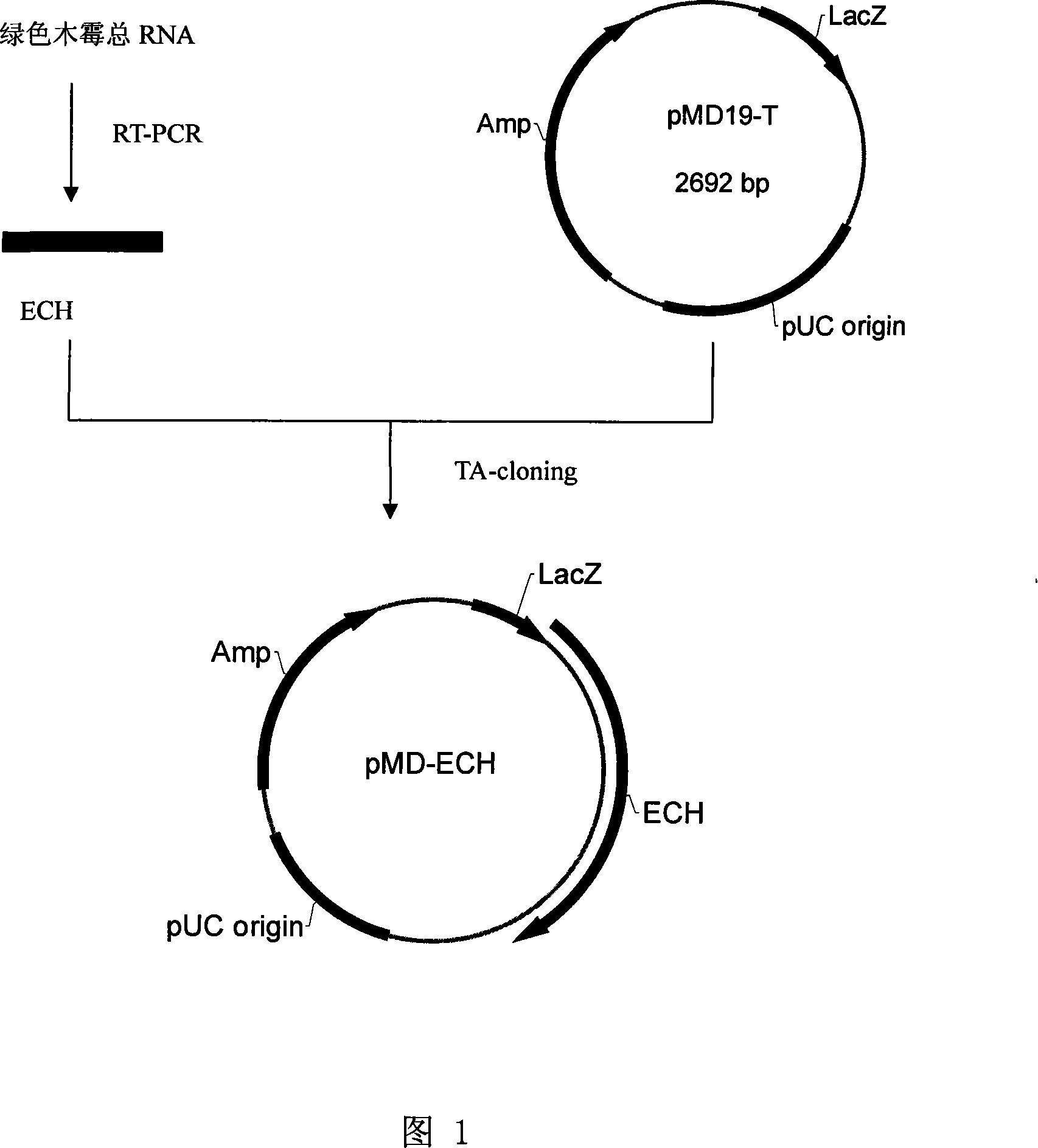
[0015] Example 1. Obtaining of Trichoderma endochitinase cDNA
[0016] Extract total RNA by Trizol method: Grind 100 mg of Trichoderma mycelium into powder in liquid nitrogen, and transfer the powder to a 1.5 mL centrifuge tube when the liquid nitrogen is not yet volatile. Use a 1mL syringe, 26-G (Ф4.5) needle to aspirate the homogenate several times to shear the genomic DNA, and then directly transfer the sample from the syringe to a 1.5mLEppendorf tube. Add 100 μL of chloroform / isoamyl alcohol (24:1), shake vigorously and mix for 30 seconds. Centrifuge at 12,000 rpm on a bench top centrifuge for 5 minutes at room temperature. Carefully transfer the supernatant to a sterile 1.5mL RNase-free centrifuge tube, add 150μL of absolute ethanol, and mix. Transfer all the above solutions to a UNIQ-10 column nested in a 2mL collection tube, leave it at room temperature for 5 minutes, and centrifuge at 8,000 rpm for 1 minute. Take out the column carefully, discard the waste liquid in th...
Embodiment 2
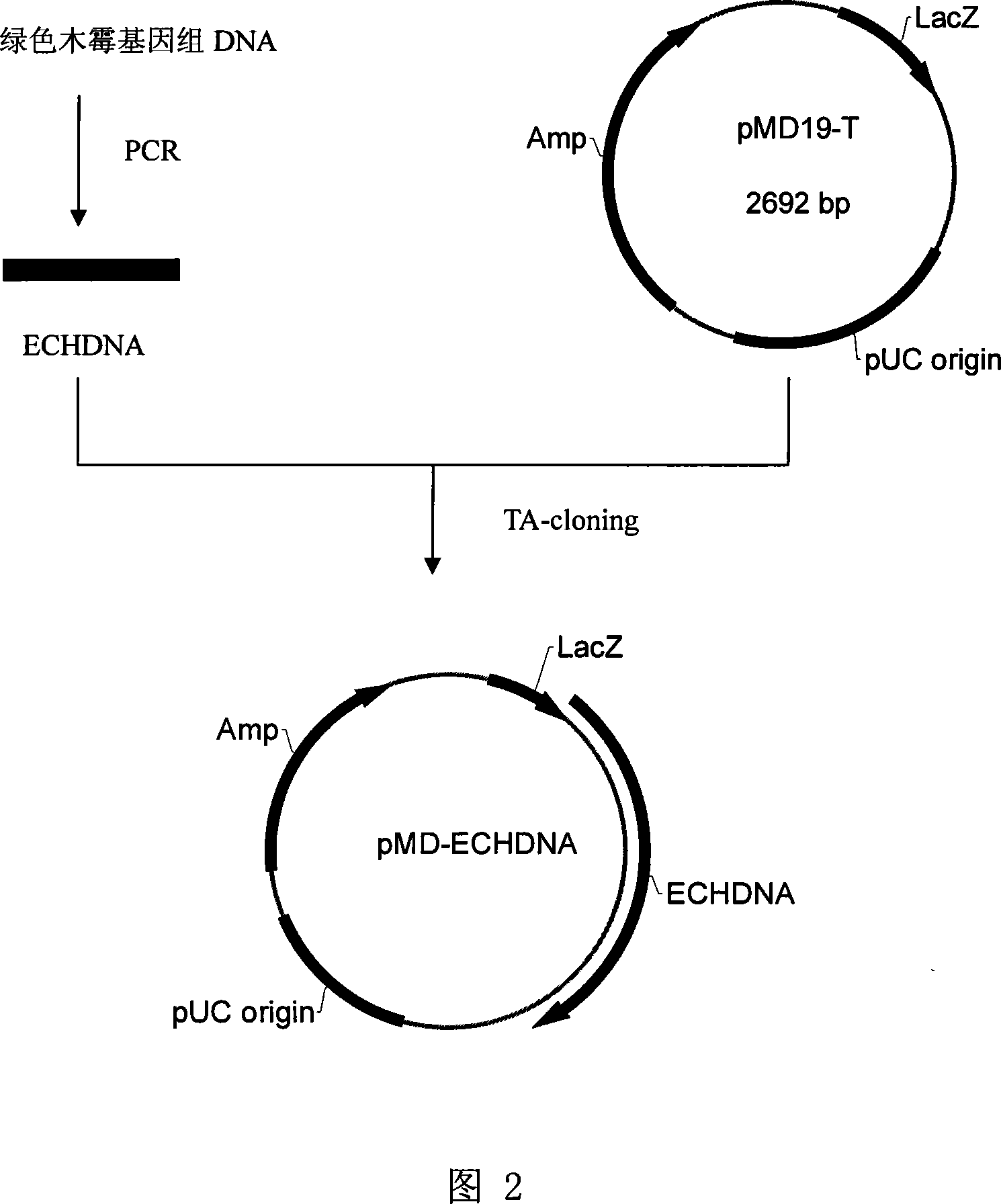
[0024] Example 2. Obtaining of Chitinase DNA from Trichoderma
[0025] Weigh 200mg (fresh weight) of Trichoderma mycelium and grind it into powder in liquid nitrogen. Use 0.5ml of solution I (100mmol / L NaCl; 30mmol / L Tris-HCl, pH7.8; 10mmol / L EDTA, pH8) .0; 10mmol / L β-mercaptoethanol) suspension, the suspension was centrifuged at 12000g for 1min to get a precipitate, and solution I (100mmol / L NaCl; 30mmol / L Tris-HCl, pH7.8; 10mmol / L EDTA, pH8 was added. 0; 10mmol / L β-mercaptoethanol) suspension and centrifuge once. The precipitate was suspended with 0.6 mL of solution II (0.1 mmol / L NaCl; 0.2 mmol / L sucrose; 10 mmol / L EDTA). Add 0.2mL lysate, shake vigorously for 1min, and place the suspension in a 55℃ water bath for 30min. Extract twice with an equal volume of phenol / chloroform / isoamyl alcohol (25:24:1) and centrifuge at 12000g for 5min. Take the supernatant and add 1 / 10 volume of 3mol / L NaAc, mix well, add 2 times volume of absolute ethanol, precipitate at room temperature f...
Embodiment 3
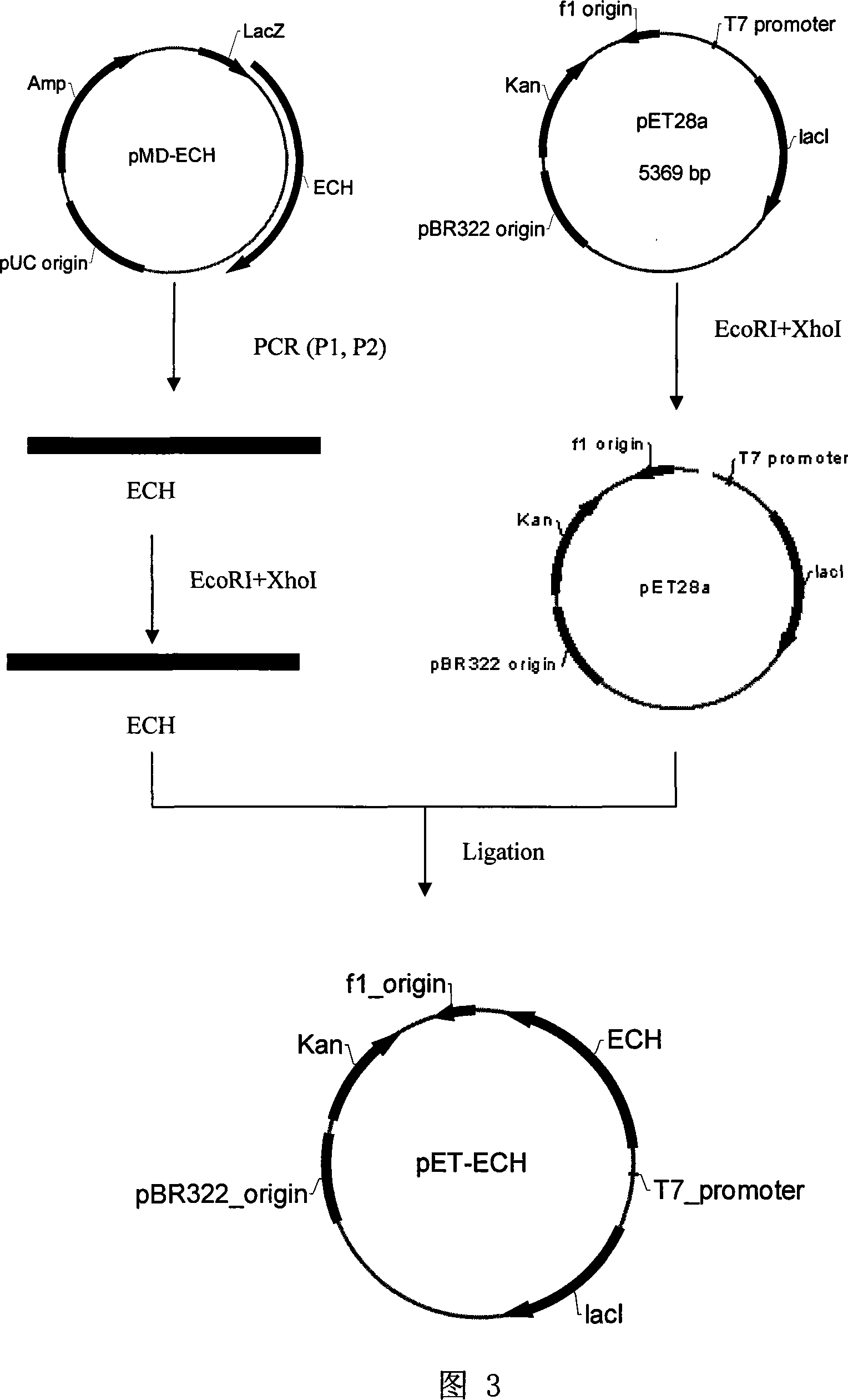
[0032] Example 3. Construction of prokaryotic expression vector of Trichoderma endochitinase cDNA
[0033] Such as image 3 Shown is the construction process of a Trichoderma endochitinase cDNA containing signal peptide prokaryotic expression vector.
[0034] The genomic DNA and cDNA sequences were compared to obtain the partial sequence of introns. Two sets of primers were designed to construct a prokaryotic expression vector containing signal peptides (primers P1, P2) and without signal peptides (primers P3, P2). The primer sequence is as follows:
[0035] P1 (SEQ ID NO.4): 5'-GACGAATTCATGTTGGGCTTCCTCGGAAAA-3'
[0036] P2 (SEQ ID NO.5): 5'-AGACTCGAGAGTTGAGACCGCTTCGGATGTT-3'
[0037] P3 (SEQ ID NO.6): 5'-CATGAATTCGCCAGCGGATATGCAAACGCC-3'
[0038] The plasmid pMD-ECH and the E. coli original expression vector pET28a (purchased from Novagen Co., Ltd., USA) were extracted by alkaline method. Using P1 and P2 as primers, using pMD-ECH as template, PCR technology was used to amplify the end...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com