Method for detecting suspension chip of multiple PCR products
A detection method and product technology, applied in the field of improvement of the conditions in the suspension chip detection method, can solve the problems of poor specificity, carcinogenicity, indistinguishable fragments of similar size, etc., and achieve the effect of flexible design
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
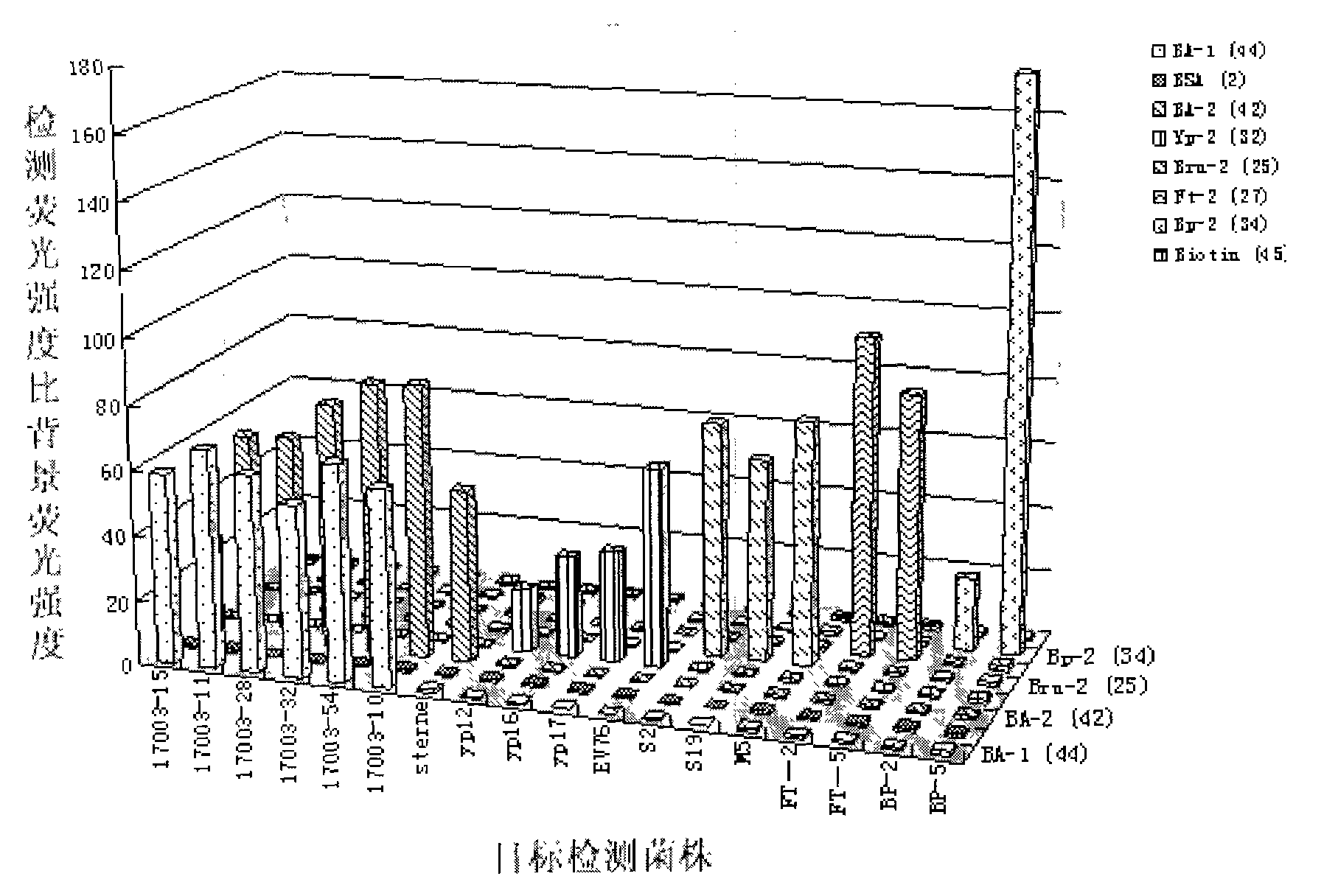
[0045] Embodiment 1: The suspension chip detection method of PCR product is used for five kinds of bioterrorism factors Bacillus anthracis, Yersinia pestis, Brucella, Francisella tularensis, Burkholderia pseudomallei six-fold PCR product detection
[0046] 1. Determine the diagnostic fragments of the above five bacteria, select capB and an identification sequence on the chromosome for anthrax, select an identification sequence on the chromosome for plague, and select the specific fragment BCSP31 of the genus Brucella for Brucella, through NCBI's GeneBank Obtain candidate gene sequences, use software to design primers, and synthesize related sequences. The downstream primers are labeled with biotin at the 5' end, see Table 1, and these primers are diluted to 10 μmol / L.
[0047] Table 1 Primer Sequence
[0048]
[0049] 2. Multiplex PCR reaction using the above primers to amplify the above five bacteria to be detected.
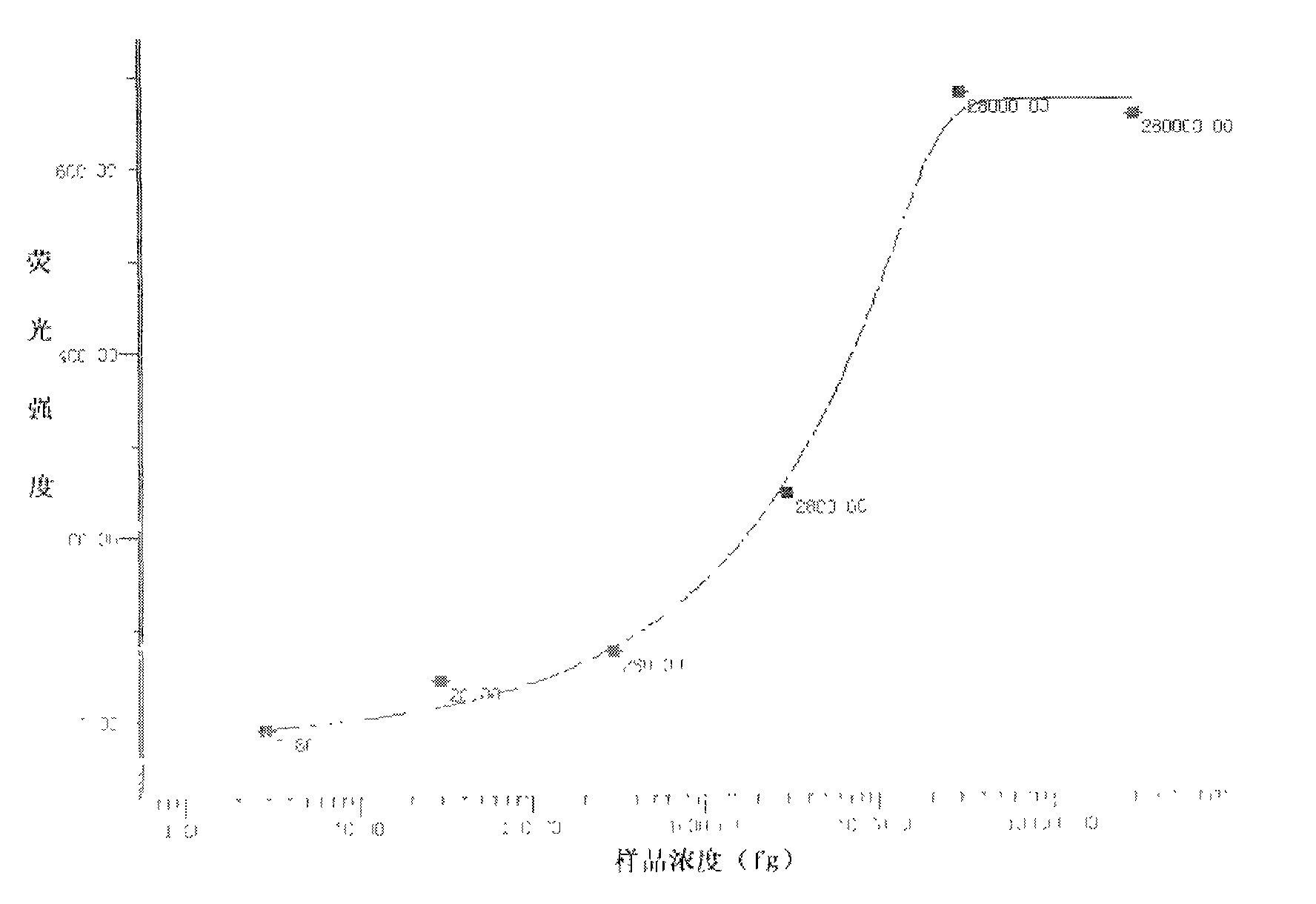
[0050] 3. According to the nucleotide sequence in the a...
Embodiment 2
[0057] Example 2: Coupling of corresponding probes to microspheres
[0058] 1) Select coded microspheres with different numbers respectively, and oscillate the microsphere suspension with a vortex shaker to mix the microspheres evenly.
[0059] 2) Take the above microspheres about 1.25×10 6 Each was transferred to a centrifuge tube, centrifuged at 14000g for 3-5min, and the supernatant was carefully aspirated.
[0060] 3) Add 50ul of 0.1mol / L 2-(n-morpholino)ethanesulfonic acid solution, shake for 20-30s, and sonicate for 20-30s to resuspend the microspheres.
[0061] 4) Dilute the synthesized oligonucleotide probe to 0.1 mmol / L with distilled water.
[0062] 5) Add 1 ul of the diluted probe to the microsphere suspension, shake and mix.
[0063] 6) Add 2.5ul of freshly prepared 10mg / mL EDC solution to the mixture of microspheres and probes, shake and mix.
[0064] 7) Wrap the centrifuge tube with aluminum foil to avoid light, oscillate on a vortex shaker at 400-600 rpm, an...
Embodiment 3
[0070] Embodiment 3: Capture and detection of the PCR product to be detected by the capture probe
[0071] i. Take 3500 each of the coupled detection probes in the hybridization solution so that the total amount is 33 μl (calculate the corresponding addition amount according to the counting result of the microspheres)
[0072] ii. Add 5-17ul of the PCR products of the five bacterial mixed templates to each tube to make the final volume 50ul, and mix well by pipetting.
[0073] iii. Denature at 95°C for 10 minutes.
[0074] iv. Hybridization at the hybridization temperature for a certain period of time.
[0075] v. Transfer to a filter plate and filter to remove unbound PCR products.
[0076] vi. Add 75ul 4ng / ul SA-PE 1×TMAC solution to each well, incubate at room temperature in the dark for 10min, and filter to remove unbound SA-PE.
[0077] vii. Add 75ul 1×TMAC solution to each well and shake to resuspend the microspheres.
[0078] viii. Check on the machine after the rea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com