mRNA difference displaying method with ordered robustness
A differential display and robust technology, applied in the field of robust and orderly differential display of mRNA, can solve the problems of dependence and little effect, and achieve the effect of credible results, high coverage and economical operation cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Study on gene expression differences between leaves and roots of one-week-old rice seedlings:
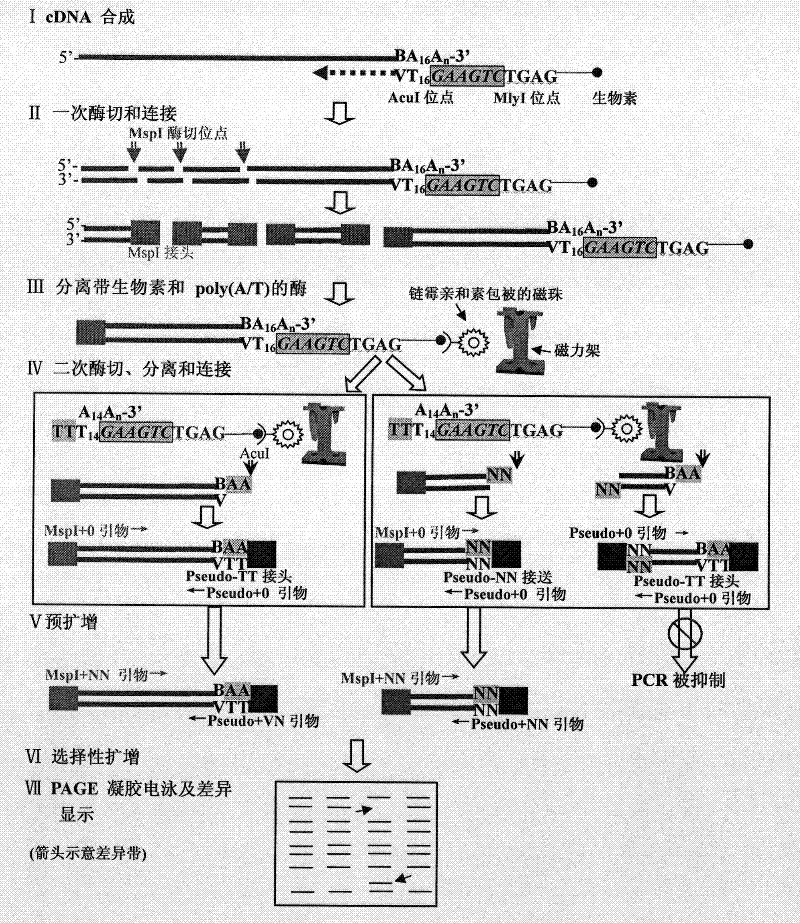
[0034] A method for robust and ordered mRNA differential display, comprising the following steps:
[0035] (1) Synthesis of double-stranded cDNA. Total RNA was extracted from middle leaves and roots of one-week-old standard japonica rice Nipponbare seedlings whose genome sequence had been determined using TRIzol reagent (Invitrogen, USA). After DNase I (Promega, USA) treatment, phenolform (volume ratio 1:1) extraction and 75% (volume percentage) ethanol precipitation, the purified total RNA was dissolved in 30 μl diethyl pyrocarbonate (DEPC) treated of water. Take an appropriate amount of purified total RNA from leaves and roots, use the designed bmaT16V as a primer (synthesized by Invitrogen, USA), and use SuperScript TM III reverse transcriptase (Invitrogen, USA) was used to synthesize the first strand of cDNA respectively. Further use RNase H, DNA polymeras...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com