Procambarus clarkia chitin peptide gene and encoded chitin peptide and application thereof
A technology of protocrayfish and crustacean, which is applied in the field of genetic engineering and can solve problems such as no reports of protocrayfish crustacean gene and the like.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Example 1: Cloning of Crayfish carapace peptide cDNA
[0069] 1) Extraction of total RNA: Total RNA was extracted by one-step method using the prior art.
[0070] 2) cDNA first-strand synthesis: 4 microliters of total RNA, plus 1 microliter of SmartF (5'-TAC GGC TGC GAGAAGACGACA GAA GGG-3') and 1 microliter of Oligoanchor R (5'-GAC CAC GCG TAT CGA TGT CGA CT 16 (A / C / G)-3'), react at 72°C for 5 minutes, then add 4 microliters of 5-fold Buffer, 1.25 microliters of dNTP, 0.625 microliters of RNase inhibitor, 1 microliter of MMLV reverse transcriptase, RNase-free 12.875 microliters of sterilized water were reacted at 42°C for 60 minutes, and the reaction was terminated at 70°C for 10 minutes.
[0071] 3) PCR reaction: chain polymerase reaction (PCR) reagents and conditions:
[0072] First mix the following reagents together:
[0073] 5 microliters (μl) of 10xTaq DNA polymerase buffer
[0074] ●Template cDNA 1μl
[0075] ●Forward primer (10mM) 1μl
[0076] ●Reverse pr...
Embodiment 2
[0096] Example 2: Construction, expression and purification of prokaryotic recombinant expression vectors
[0097] (1) According to the sequence of Procambarus clarkii crustacean peptide (remove signal peptide sequence) and the cloning site of expression vector pPET30a (Novagen Company), design primers:
[0098] Cru-pET F:5′TAC TCA GAA TTC CAC CTT AAA CGA CCC AAG CC 3′(EcoRI)
[0099] Cru-pET R:5′TAC TCA CTC GAG CTA ATT GAT GTA AAA GTG TGC 3′(XhoI)
[0100] The present invention selects the EcoR I and XhoI restriction sites of the pET30a cloning site. Therefore, when designing primers, an EcoR I restriction site is introduced into the upstream primer, and an Xho I restriction site is introduced into the downstream primer.
[0101] (2) Gene amplification, cloning and recombinant plasmid screening
[0102] Using pMD-18T-Lec as template, carry out PCR reaction with the above primers, the amplification conditions are: 94°C, 2min pre-denaturation; 94°C, 30s, 56°C, 45s, 72°C, ...
Embodiment 3
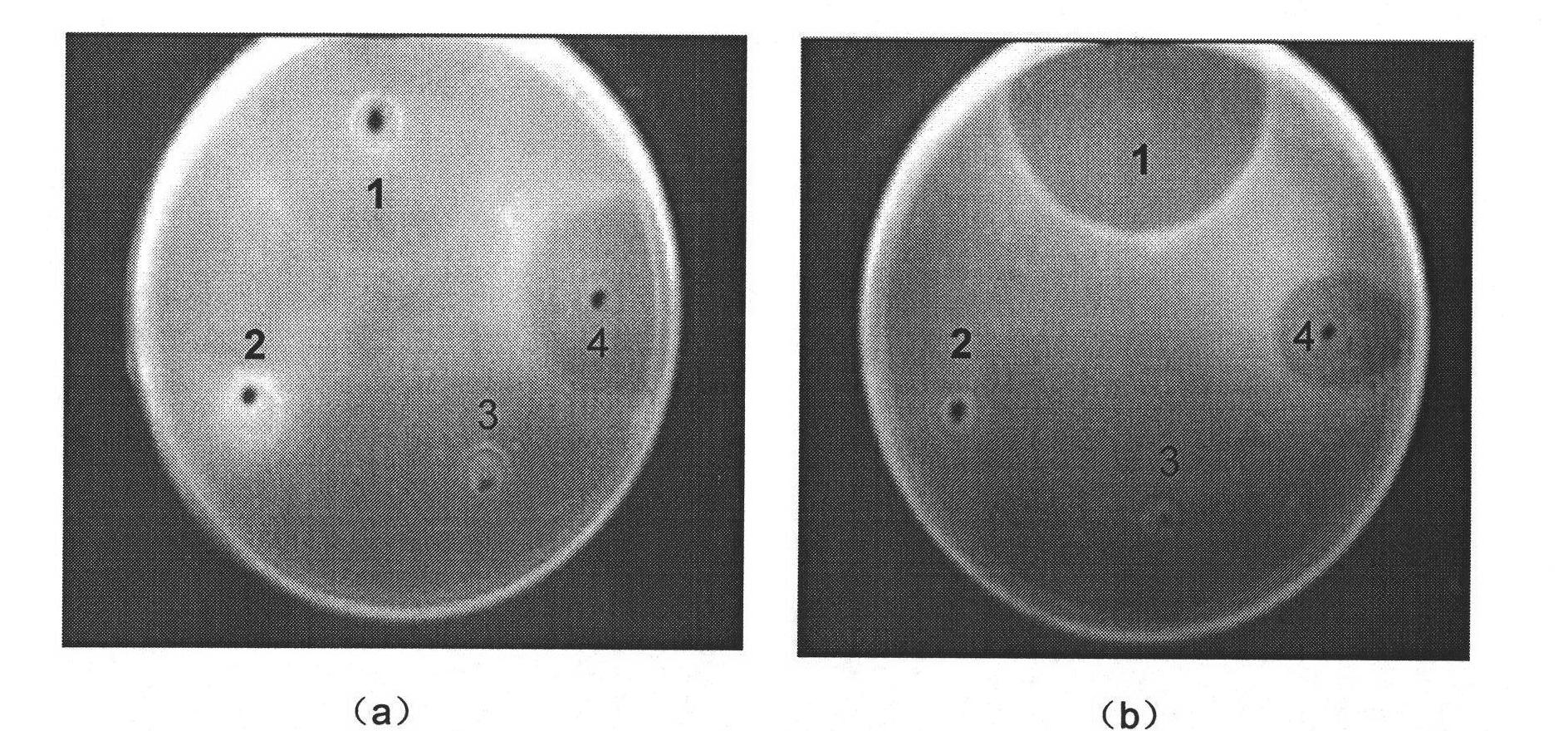
[0113] Example 3: Recombinant expression and antibacterial activity determination of chitin in Pichia pastoris
[0114] (1) Construction of pPIC9K (Invetrogen Company) recombinant expression vector
[0115] The present invention selects the EcoR I and Not I restriction sites of the pPIC9K cloning site. Therefore, when designing the primers, the EcoR I restriction site is introduced into the upstream primer, and the Not I restriction site is introduced into the downstream primer:
[0116] Cru-pPIC F:5′TAC TCA GAA TTC CAC CTT AAA CGA CCC AAG CC 3′(EcoRI)
[0117] Cru-pPIC R:5′TAC TCA GCGGCCGC CTAs ATG ATG ATG ATG ATG ATG ATG ATG ATG ATT GAT GTA AAA GTG TGC 3′ (NotI)
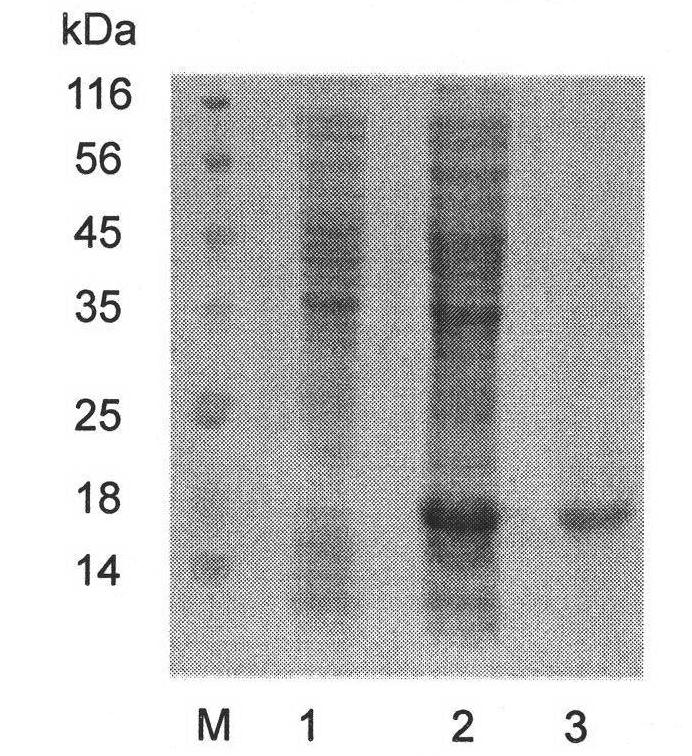
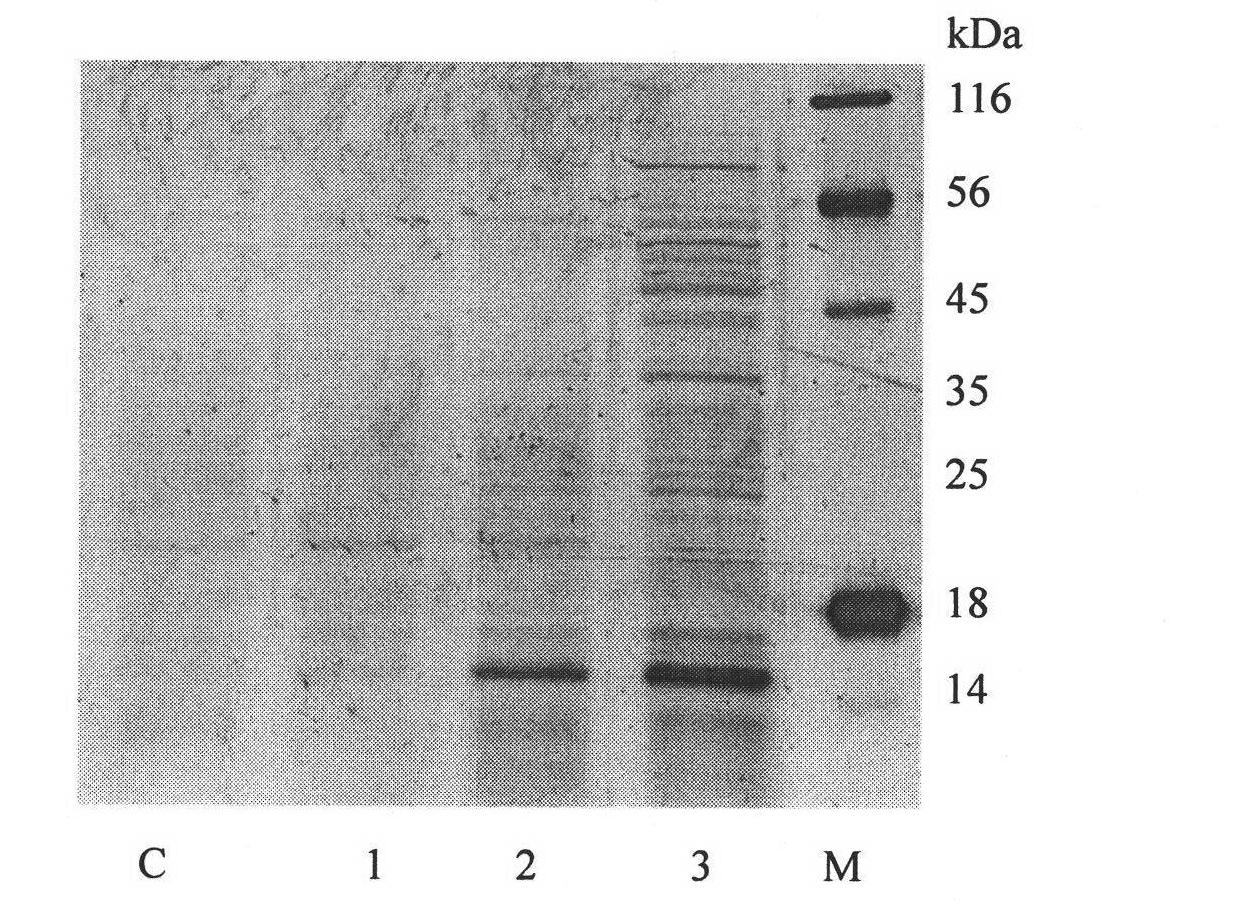
[0118] Using cDNA of blood cells of Procambarus clarkii as template, the sequence of mature peptide of crustacean was amplified by conventional PCR method, and the PCR amplification product was purified. Then the PCR amplified fragments of pPIC9K and Crustin were digested with EcoR I and Not I. The diges...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com