Codon-optimized H3HA/XJ3-07 gene and nucleic acid vaccine thereof
A technology of codon optimization and nucleic acid vaccine, which is applied in the field of H3HA/XJ3-07 gene and its nucleic acid vaccine, can solve the problems of gene cloning difficult to express effectively, stimulating host immune system, and low immunogenicity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
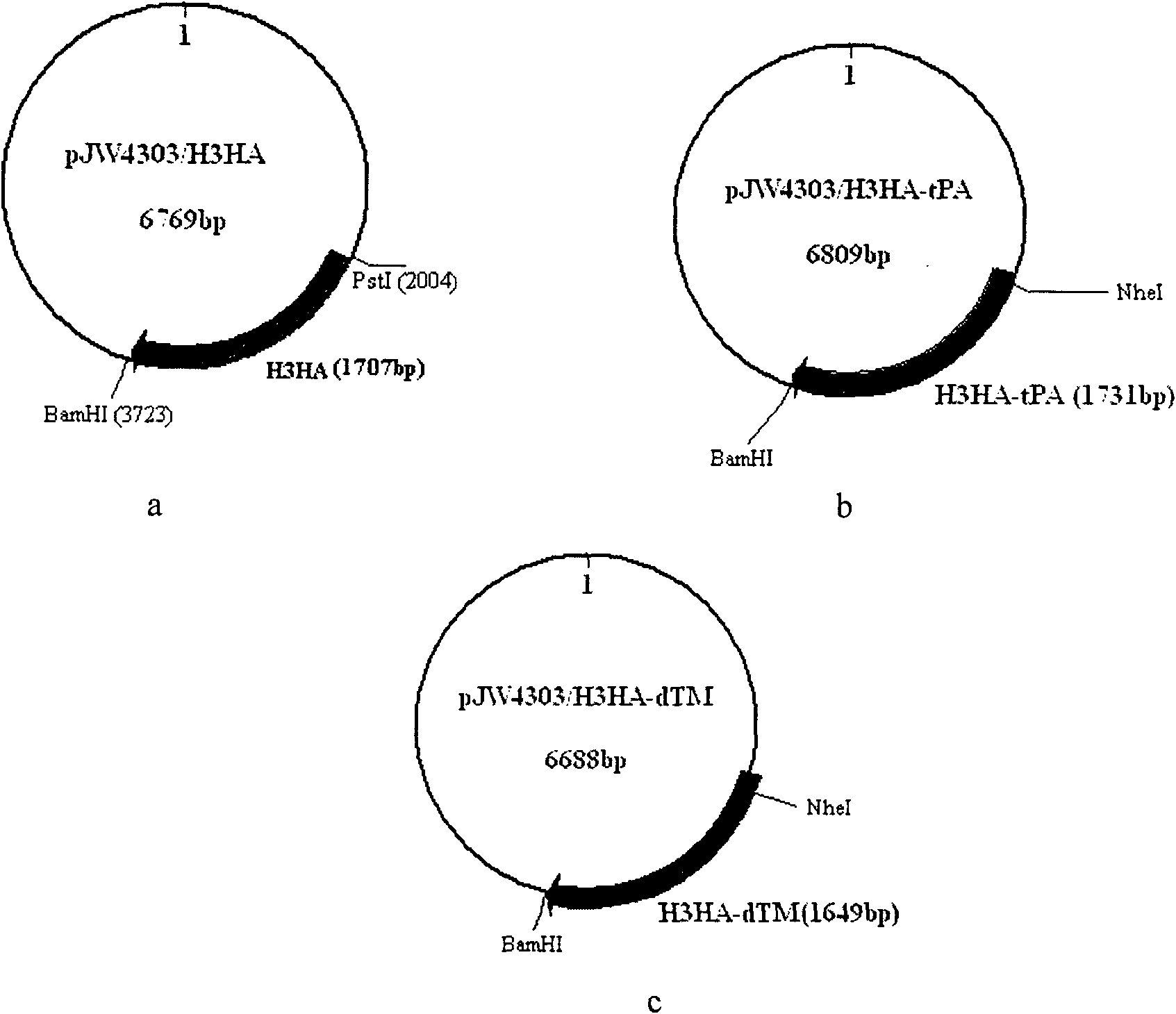
[0040] Example 1. Construction and Identification of Nucleic Acid Vaccine pJW4303 / H3HA
[0041] 1.1 Preparation of Competent Escherichia coli HB101
[0042] 1.1.1 Thaw Escherichia coli HB101 preserved in glycerol on ice, inoculate 5ul into 5ml LB solution, place in a constant temperature incubator at 37°C, 180rpm, and shake the bacteria overnight.
[0043] 1.1.2 Under sterile conditions, take 1ml of the overnight culture solution, inoculate it into 200ml of fresh LB solution, place it in a constant temperature incubator at 37°C, 180rpm, and shake for 2 hours.
[0044] 1.1.3 Cool the bacteria in the previous step on ice for 15 minutes.
[0045] 1.1.4 Aseptically dispense into 50ml centrifuge tubes (autoclaved), centrifuge at 2500rpm at 4°C for 15min.
[0046] 1.1.5 Add cold 0.1M Cacl 2 100ml (Cacl 2 have been autoclaved), resuspend the bacteria, and incubate on ice for 30 min.
[0047] 1.1.6 Aseptically dispense into 50ml centrifuge tubes (autoclaved), centrifuge at 2500r...
Embodiment 2
[0085] Example 2. Construction and Identification of Nucleic Acid Vaccine pJW4303 / H3HA-tPA
[0086] 2.1 Design PCR primers, respectively: upstream primer SEQ ID N0.4, downstream primer SEQ ID N0.5; use the recombinant plasmid pGA15 / H3HA as a template to amplify the GCTAGC (NheI) and GGATCC (BamHI) H3HA gene fragment (SEQ ID NO.2) that does not contain the natural signal peptide gene at the restriction site. The reaction conditions are: 94°C for 4min, 94°C for 1min, 56°C for 1min, 72°C for 1.5min, a total of 25 cycles, and 72°C for 7min.
[0087] 2.2 Take 2ul of the PCR product and run 1% agarose gel electrophoresis for identification.
[0088] 2.3 Identify the correct PCR product with DNA gel recovery kit (E.Z.N.A. TM Gel Extraction Kit (50), OMEGABIO-Tek Company) recovered and purified H3HA gene fragments. The specific steps of glue recovery are as 1.4 operation process.
[0089] 2.4 Purified PCR product and vector pJW4303 were double digested with NheI and BamHI resp...
Embodiment 3
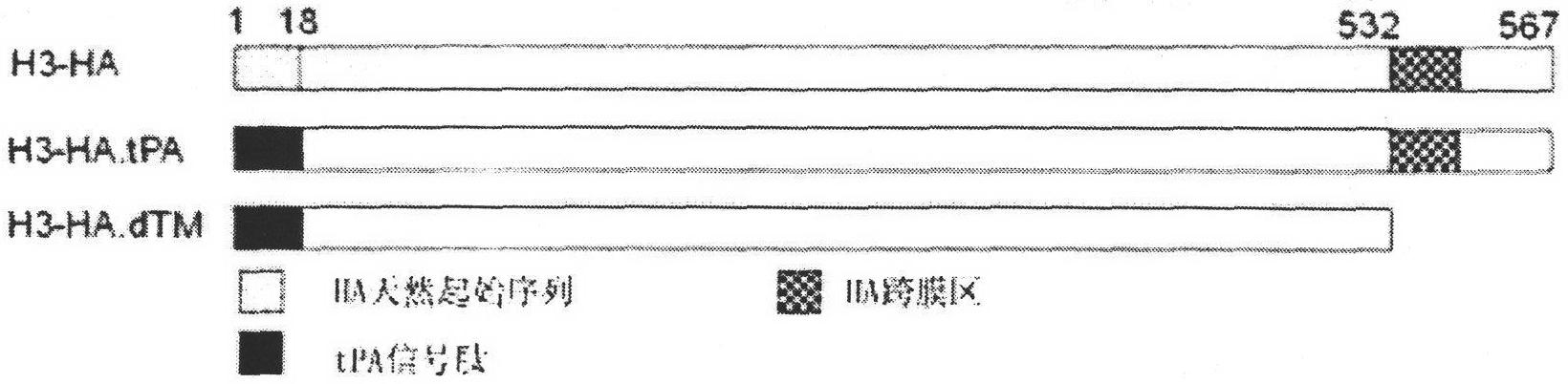
[0117] See the physical map of pJW4303 / H3HA-tPA nucleic acid vaccine plasmid figure 2 b. The sequence located between the NheI and BamHI endonuclease sites is the H3HA gene. For enzyme digestion results, see Figure 4 Lane1, lane5, Lane6, lane7. After Nhe+BamHI double digestion, the target gene fragment H3HA-tPA was released, the length of which was slightly smaller than 1700bp, as expected, and the pJW4303 / H3HA-tPA nucleic acid vaccine was successfully constructed. Example 3. Construction and Identification of pJW4303 / H3HA-dTM Nucleic Acid Vaccine
[0118] 3.1 Design two pairs of PCR primers, both using the recombinant plasmid Pga15 / H3HA as a template, using the upstream primer SEQ ID NO.4 and the downstream primer SEQ ID NO.6 to amplify the GCTAGC (NheI) and GGATCC H3HA gene fragment (SEQ ID NO.3) of (BamHI) restriction site. The PCR reaction conditions are the same as those in 2.1. The gene fragment is a gene fragment that only encodes the extracellular domain part...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com