DNA for high-level expression of N-acyl homoserine lactonase in yeasts and engineering bacteria constructed thereby
A technology of acyl homoserine lactone and DNA molecules, applied in the direction of genetic engineering, recombinant DNA technology, microorganism-based methods, etc., to achieve the effects of reducing production costs, improving safety and effectiveness, and high expression ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1, Optimization of N-acyl homoserine lactonase gene codons
[0051] 1. Optimization of codons of homoserine lactonase gene
[0052] The N-acyl homoserine lactonase gene aiiaB546 (shown in sequence 1 of the sequence listing) is codon-optimized, and the optimized gene aiiaB546M is shown in sequence 2 of the sequence listing. For comparison of gene aiiaB546 and optimized gene aiiaB546M see figure 1 .
[0053] 2. Construction of recombinant plasmid aiiaB546 / pUC57
[0054] 1. Artificially synthesize the aiiaB546 gene shown in sequence 1 of the sequence listing.
[0055] 2. Digest the gene synthesized in step 1 with restriction endonucleases EcoR I and Not I, and recover the digested product.
[0056] 3. Digest the pUC57 vector with restriction enzymes EcoR I and Not I to recover the vector backbone.
[0057] 4. Ligate the digestion product of step 2 with the vector backbone of step 3 to obtain a ligation product.
[0058] 5. The ligation product was sequenced, ...
Embodiment 2
[0065] Embodiment 2, construction of recombinant expression vector
[0066] 1. Construction of pPIC9-aiiaB546M
[0067] 1. Extract recombinant plasmid aiiaB546M / pUC57 and plasmid pPIC9 respectively
[0068] Plasmids were extracted using a plasmid mini-extraction kit from Beijing Tiangen Biotechnology Co., Ltd., and detected by 1.2% agarose gel electrophoresis (electrophoresis buffer 1×TAE, voltage 1-5 V / cm, time about 30 min). The electropherogram of the recombinant plasmid aiiaB546M / pUC57 is shown in figure 2 .
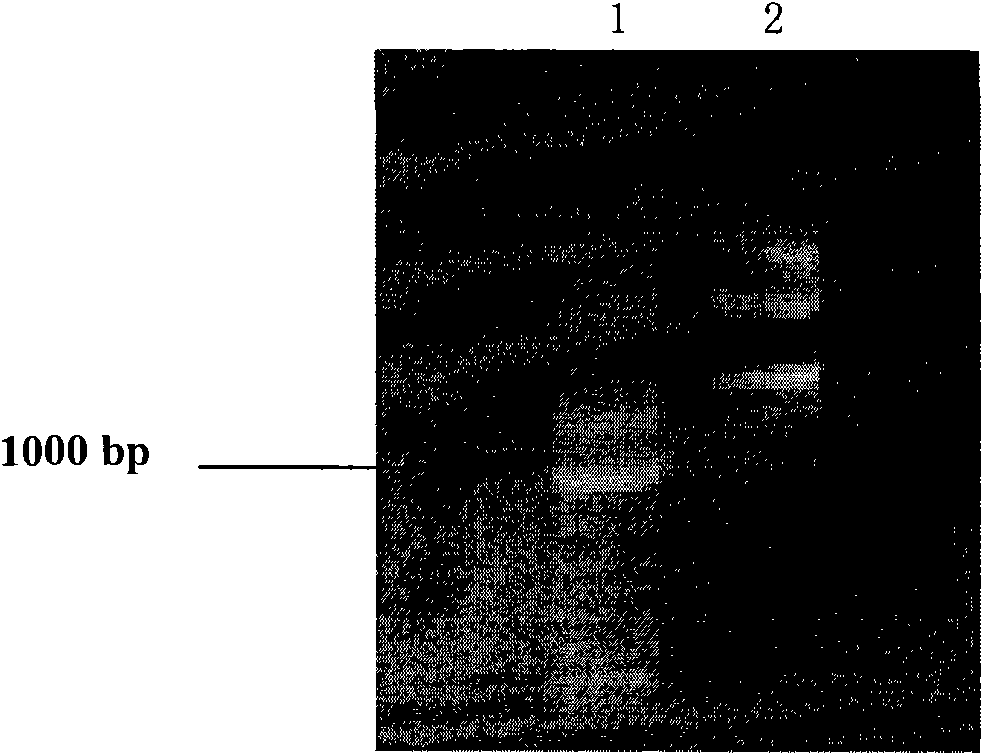
[0069] 2. Digest plasmid pPIC9 with restriction endonucleases EcoR I and Not I to recover the vector backbone. 1.0% agarose gel electrophoresis detection, see electrophoresis image 3 .
[0070] 3. Digest the recombinant plasmid aiiaB546M / pUC57 with restriction endonucleases EcoR I and Not I, and recover a DNA fragment (aiiaB546M) of about 750 bp. 1.0% agarose gel electrophoresis detection, see electrophoresis image 3 .
[0071] 4. Ligate the vector backbon...
Embodiment 3
[0083] Embodiment 3, the preparation of recombinant bacteria
[0084] 1. Preparation and screening of recombinant bacteria with aiiaB546M gene
[0085] 1. Mass extraction of plasmid DNA
[0086] ① Take the Escherichia coli Trans1 containing the recombinant plasmid pPIC9-aiiaB546M obtained in Example 2 and inoculate it in 50 mL LB medium containing 100 μg / mL ampicillin, and cultivate overnight at 37°C with shaking;
[0087] ② Take 50 mL of the overnight culture solution, centrifuge at 10,000 rpm, 4°C for 5 minutes, discard the supernatant, invert the centrifuge tube on absorbent paper, and absorb the excess liquid;
[0088] ③Resuspend the precipitate in 2mL of solution I, shake vigorously, and suspend fully; add 4mL of freshly prepared solution II, mix upside down, and place on ice for 4min; add 3mL of solution III, mix upside down, place on ice for 5min, 13,000rpm , Centrifuge at 4°C for 10 minutes;
[0089] ④Remove the supernatant and filter it with lens-cleaning paper; ad...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com