Method, gene chip and detection reagent kit for detecting common cow pathogenic escherichia coli
A technology of Escherichia coli and gene chip, which is applied in the field of detection of Escherichia coli serotypes that cause bovine septicemia and diarrhea, can solve the problems of limited information in one detection, inapplicability of a large number of samples, etc., and achieve high accuracy and easy operation , highly reproducible effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 Design and preparation of probe
[0045] 1. Sequence acquisition:
[0046] (1) Obtaining the bacterial 16s rDNA sequence: Download the 16s rDNA sequence from the GenBank public database.
[0047] (2) Obtaining the O antigen synthesis gene cluster sequence of Escherichia coli O8, O9, O15, O26, O86: The 5 O antigen serotype E. coli O antigen synthesis gene cluster sequences downloaded from the GenBank public database.
[0048] (3) Obtaining the O antigen-specific gene sequences of E. coli O35, O78, O101, O115 and O119: The O antigen synthesis gene cluster sequences of E. coli O35, O78, O101, O115 and O119 have been sequenced by the inventor's unit.
[0049] 2. Probe design:
[0050] Import the above sequence into OligoArray2.0 software, and design the probe according to the instructions of the software.
[0051] In a preferred embodiment of the present invention, 43 probes designed by OligoArray 2.0 software are selected, and probes are screened through hybridization exper...
Embodiment 2
[0058] Example 2 Design and preparation of primers
[0059] 1. Sequence acquisition: same as the sequence of the designed probe.
[0060] 2. Design primers:
[0061] Import the above sequence into the primer design software Primer Premier 5.0 software, and refer to the software's instructions for primer design.
[0062] In the preferred embodiment of the present invention, the main Escherichia coli serotypes (O8, O9, O15, O26, O35, O78, O86) that both contain probes and are suitable for Multiplex PCR are selected in the preferred embodiment of the present invention. , O101, O115 and O119) 11 pairs of primers, in order to adapt to Multiplex PCR, through the preliminary screening of bioinformatics and a large number of Multiplex PCR experiments, the suitable primers as shown in Table 2 were selected.
[0063] Table 2 The primer sequences used in PCR amplification for detection of 10 common bovine pathogenic E. coli
[0064]
[0065]
[0066] 3. Primer synthesis: commission the primer syn...
Embodiment 3
[0067] Example 3 Preparation of gene chip-chip spotting
[0068] 1. Dissolving the probes: the probes synthesized in Example 1 were respectively dissolved in 50% DMSO solution, and diluted to make the final concentration of the probe reach 1 μg / μl.
[0069] 2. Add plate: add the dissolved probe to the corresponding position of the 384-well plate, 10μl per well.
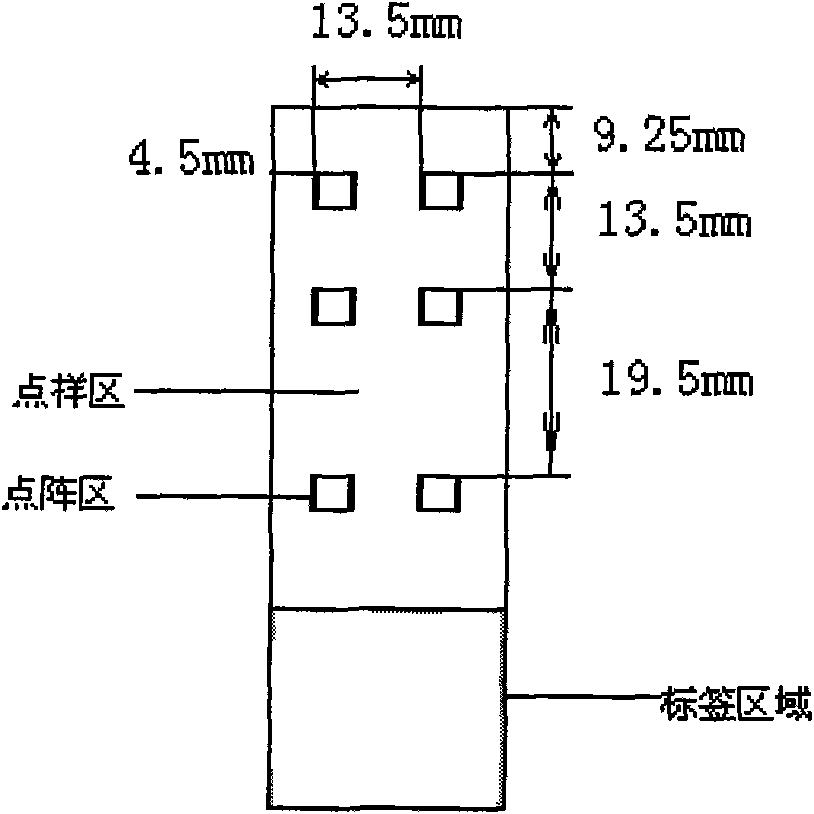
[0070] 3. Pointing: Will be as figure 1 The shown 57.5mm×25.5mm×1mm (L×W×H) clean aldehyde-based glass slide (CEL Associates, Inc.) is placed on the stage of the chip spotter (Spotarray 72), and SpotArray is used Control software (Tele chem smp3 stealty pin), run the program, press figure 2 The arrangement shown is dotted in a 4.5mm×4.5mm spot area on the aldehyde-based glass slide, forming a medium and low density DNA micro-matrix. The six dot matrix areas on the glass slide have the same array arrangement. The size of the dot matrix area is 3mm×2.25mm, the dot pitch in the dot matrix is 250μm, the matrix: 12×9, 12×250...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com