Detection method for distinguishing non-pneumophilia legionella and legionella pneumophilia and kit
A technology for Legionella pneumophila and Legionella pneumophila, which is applied in microorganism-based methods, biochemical equipment and methods, and determination/inspection of microorganisms, etc., can solve problems such as inability to judge results and inconspicuous melting curve characteristics of Legionella pneumophila. , to achieve stable and reliable results, easy and fast operation, and avoid misdiagnosis.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
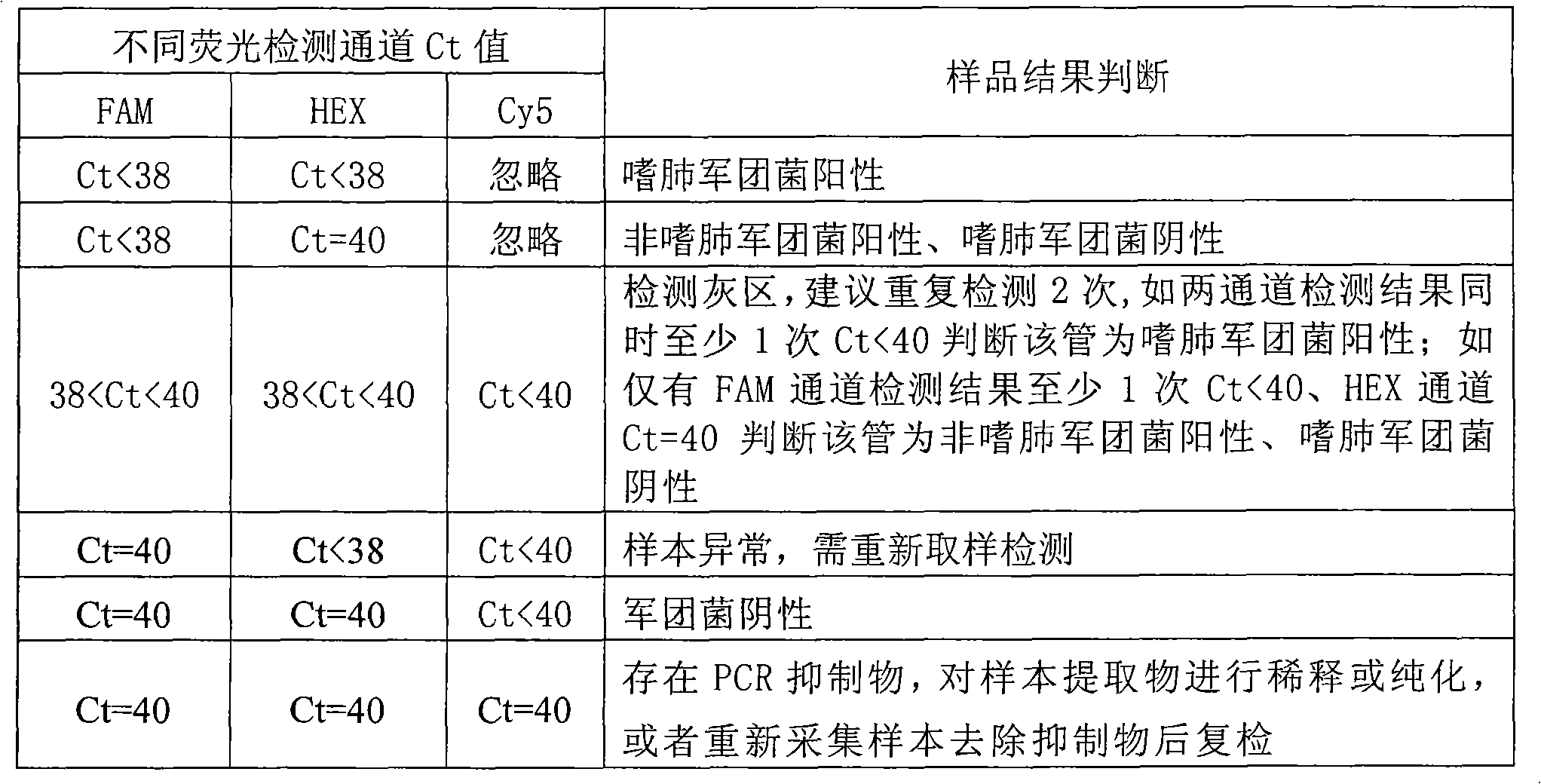
Image
Examples
Embodiment 1
[0051] Embodiment 1: the preparation of Legionella nucleic acid detection kit (PCR-fluorescent probe method)
[0052] (1) Primer probe synthesis
[0053] Design the primer probe according to the target sequence SEQ ID No.1, and entrust a commercial sequence synthesis company (Shanghai Sangon Bioengineering Technology Service Co., Ltd.) to synthesize:
[0054] The upstream primer is 5'-TgTgAAATTCCTgggCTTAAC-3', as shown in SEQ ID No.2;
[0055] The downstream primer is 5'-TTCgTgCCTCAgTgTCAg-3', as shown in SEQ ID No.3;
[0056] The Legionella fluorescent probe is 5'-CAggTAgCCgCCTTCgCC-3', the 5' end is labeled with FAM (6-carboxyfluorescein), and the 3' end is labeled with BHQ (Black Hole Quencher)), as shown in SEQ ID No.4.
[0057] The fluorescent probe for Legionella pneumophila is 5'-TgggACggTCAgATAATACTggTT-3' (5' end-labeled HEX (6-carboxyhexachlorofluorescein) or JOE (6-carboxy-4', 5'-dichloro-2', 7 '-dimethoxyfluorescein), VIC fluorophore, 3' end labeled BHQ), as sho...
Embodiment 2
[0077] Embodiment 2: the application of Legionella nucleic acid detection kit (PCR-fluorescent probe method)
[0078] (1) Sample processing
[0079] Take the collected water sample, filter it through a filter membrane (pore size 0.22-0.45 μm), put it in a centrifuge tube after elution, centrifuge at 13,000 rpm for 6 minutes, and carefully discard the supernatant (in order to prevent the precipitation from being sucked, 5 μl residual liquid). Add 50 μl of nucleic acid extraction solution to the pellet, vortex to mix, keep at 100°C for 10 minutes, centrifuge at 13,000 rpm for 6 minutes, and take the supernatant for fluorescent PCR amplification detection. The negative and positive controls of the kit are processed in the same way as the sample eluent.
[0080] (2) Amplification detection
[0081] According to the method in the summary of the invention, each component of the unsealed kit was freeze-thawed and oscillated, and then the internal reference was added to the PCR buf...
Embodiment 3
[0084]Embodiment 3: the application of Legionella nucleic acid detection kit (PCR-fluorescent probe method)
[0085] (1) Sample processing
[0086] Take clinically collected throat swabs, sputum, and bronchoalveolar lavage fluid (the throat swabs are first rinsed with 1ml of normal saline, and the sputum is first mixed and liquefied with an equal volume of 4% NaOH), and the throat swab rinse fluid, liquefied phlegm The solution and bronchoalveolar lavage solution were placed in centrifuge tubes respectively, the sample solution was centrifuged at 13,000 rpm for 6 minutes, and the supernatant was carefully discarded (5-10 μl of the residual solution can be retained to prevent the precipitation from being sucked away). Add 50 μl of nucleic acid extraction solution to the pellet, vortex to mix, keep at 100°C for 10 minutes, centrifuge at 13,000 rpm for 6 minutes, and take the supernatant for fluorescent PCR amplification detection. The negative and positive controls of the kit a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com