Fluorescent quantitative PCR method for simultaneous detection of streptococcus suis subtype 2 (SS2) extracellular protein factor and hemolysin gene
A fluorescent quantitative, extracellular protein technology, applied in the field of nucleic acid detection, to achieve the effects of short detection time, strong reagent versatility, avoiding pollution and dispersing toxins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] According to the EF and SLY gene sequences of Streptococcus suis type 2 published in GenBank, the conserved sequences were selected through gene sequence comparison, and the GeneBank accession numbers of the selected EF and SLY public sequences were DQ417121.1 and DQ443530.1, respectively. EF and SLY primers were designed by Primer PremierS 5.0 and Oligo 6 software, and the primers were synthesized by Shanghai Sangon Biotechnology Company.
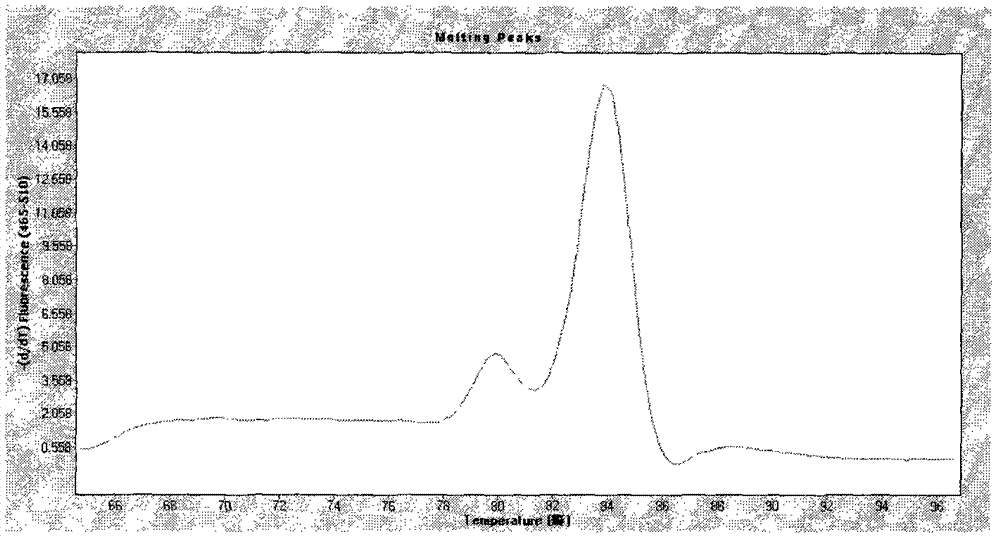
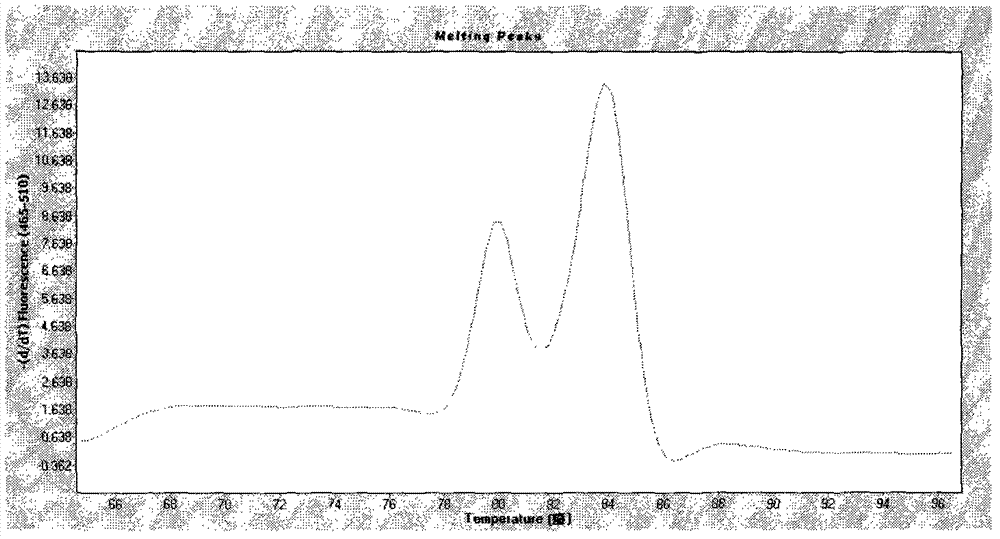
[0030] The upstream primer of EF is: 5'-GAAGAAGAACCCAAGGAACC-3'; the downstream primer of EF is: 5'-ACATTCTGACCACTCGCATC-3'. The size of the amplified EF fragment is 158bp, and the Tm value of the product is 84°C.
[0031] The upstream primer of SLY is: 5'-TTCCGATTTCGTATTCAACC-3'; the downstream primer of SLY is: 5'-AACTGTTTCCACCACTCCC-3'. The size of the amplified SLY fragment is 338bp, and the Tm value of the product is 80°C.
[0032] The extraction of embodiment 2 bacterial DNA
Embodiment 2
[0033] Inoculate sterilized Todd-Hewitt broth medium with a single colony of Streptococcus suis type 2 HA9801 strain, culture overnight at 37°C, take 1 mL of the bacterial solution, centrifuge at 12,000 rpm for 2 min, and resuspend the precipitate with 567 μL of TE buffer (pH 8.0). Add 30 μL of 10% SDS and 3 μL of 20 mg / mL proteinase K, mix well, bathe in 37°C water for 1 hour, add 100 μL of 5mol / L NaCl, mix well, then add 80 μL CTAB / NaCl solution, mix well, bathe in water at 65°C for 10 min , add an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1) and mix evenly, centrifuge at 12000rpm for 10min, absorb the supernatant and add an equal volume of chloroform:isoamylalcohol (24:1) and mix evenly, centrifuge at 12000rpm for 10min, Aspirate the supernatant, add 0.6 volume of isopropanol, place at -20°C for 20 minutes to fully precipitate the DNA, centrifuge at 12,000 rpm for 10 minutes, wash the precipitate twice with 70% ethanol, and then dry it at room temperature to...
Embodiment 3
[0036] Fluorescent PCR reaction system:
[0037] The fluorescent quantitative PCR reaction system of the present invention is a 20ul system, operated on ice, and the following reagents are added in order:
[0038]
[0039] Among them, Master Mix is SYBR Green I dye, polymerase, dNTP, Mg 2+ , PCR buffer mixture. Fluorescent PCR reaction conditions:
[0040] Pre-denaturation: 95°C, 5min
[0041] Fluorescent PCR amplification for 40-45 cycles: denaturation at 95°C for 10s; annealing at 55°C for 20s; extension at 72°C for 30s
[0042] Melting curve reaction program: 95°C, 5s; 65°C, 1min; 97°C, continuous; heating rate 2.2°C / s, start collecting fluorescence signals continuously at 65°C until 97°C.
[0043] Cooling: 40°C, 10s
[0044] Fluorescent PCR reaction criteria:
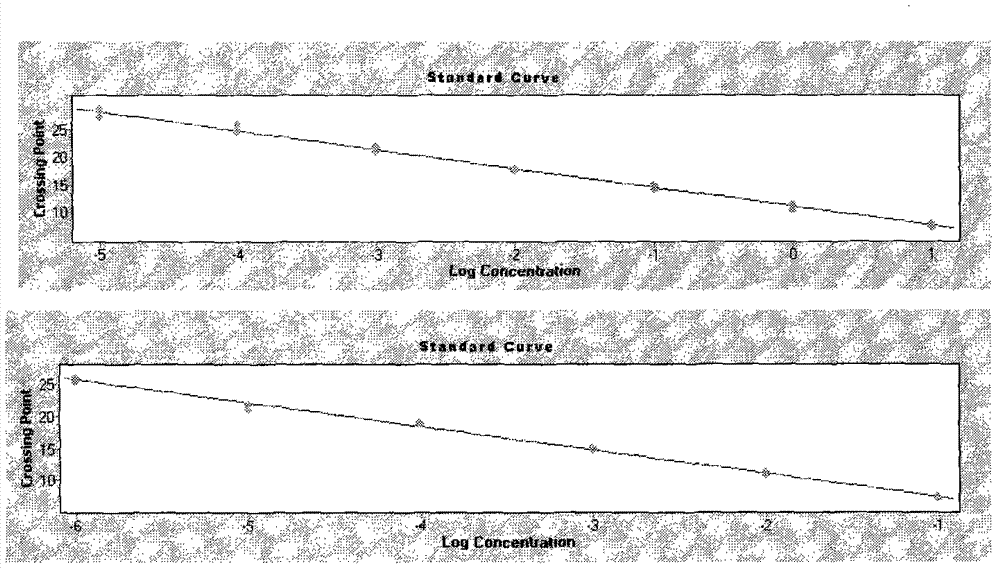
[0045] According to the Ct value and Tm value, the detection results of Streptococcus suis type 2 extracellular protein factor and hemolysin gene were judged.
[0046] The criteria for judging the amplif...
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com