Fluorescent quantitative polymerase chain reaction (PCR) kit for Acinetobacter baunannii/calcoaceticus complex OXA-23 mutant gene
A technology for OXA-23 and mutant genes, applied in the field of drug resistance gene detection kits, can solve the problems of infection control and clinical treatment difficulties, endangering the lives of patients, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1: Determination of the new sequence of the drug-resistant gene OXA-23 of Baumann's complex Acinetobacter calcium acetate
[0019] 1. Source of strains: From October 2009 to May 2011, we collected 32 strains of Acinetobacter calcoacetate baumannii complex that were highly resistant to antibiotics (especially resistant to imipenem). The 32 specimens were derived from the patient's sputum.
[0020] 2. Design of OXA-23 primers: the present invention designs a pair of specific primers for class D β-lactamase resistance gene OXA-23. The sequences of the primers are shown in SEQ ID NO: 1 and SEQ ID NO: 2.
[0021] 3. Amplification of OXA-23 gene: with above-mentioned 32 strains of Baumann's complex Acinetobacter calcium acetate lysate as template, carry out PCR amplification, amplification system is (25ul system): template DNA (bacteria lysate) 2ul, 2ul each of primers 1 and 2, TaqE: 0.25ul (0.5U), 10XBuffer 2.5ul, dNTP 2ul, with ddH 2 O make up to 25ul. The reacti...
Embodiment 2
[0029] Example 2: Preparation of a real-time fluorescent quantitative PCR kit for detecting the new sequence of OXA-23
[0030] 1. Primer design and probe synthesis
[0031] The sequence of SEQ ID NO: 3 obtained according to Example 1 above. Probes and primers (synthesized by Shanghai Jikang Company) were designed using ABI primer Express 2.0 software. See SEQ ID NO: 4 for the sequence of the upstream primer, see SEQ ID NO: 5 for the sequence of the downstream primer, and see SEQ ID NO: 6 for the sequence of the probe. The 5' end of the probe is labeled with a fluorescent chromophoric group FAM, and the 3' end is connected with a fluorescent quencher group BHQ.
[0032] 2. Standard DNA template preparation
[0033]Using the upstream and downstream primers described in the previous step, a 200bp fragment was amplified using the above-mentioned AB37 strain as a template, and the PCR product was purified (QIAgen, German) and connected to the PGM-T cloning vector and transforme...
Embodiment 3
[0039] Embodiment 3 in vitro detection experiment
[0040] 1. Sensitivity test
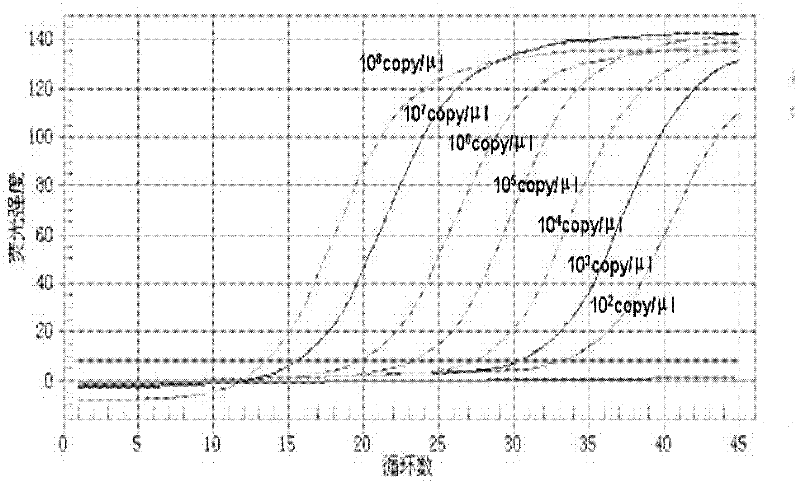
[0041] The standard DNA template was serially diluted 10 times, so that the concentration range of the standard plasmid was 10 8 ~10 2 copies / μl, as the standard of the fluorescence quantification kit. Fluorescent quantitative PCR reaction conditions, 25 μl reaction system: 12.5 μl 2×TaqMan Universal PCR Master Mix (ABI Company, USA), 10 nM each of the upstream and downstream primers (the sequences of the upper and lower primers are Sequence NO.4 and Sequence NO.5, respectively), 400nM probe (probe sequence is Sequence NO.6), 2μl gradient diluted standard DNA template, add ddH 2 0 to 25 μl. The reaction program was pre-denaturation at 95°C for 10 min, followed by 45 cycles: 95°C for 40 s, 60°C for 1 min. Sterile water was used as a negative control.
[0042] See the test results figure 1 , Sensitive analysis shows that a minimum of 100copies / reaction can be detected. This experiment was re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com