Pretreatment method for directly detecting infection urine pathogen by MALDI-TOF MS
A technology for pathogenic bacteria and urine, applied in the field of microbiology, can solve problems such as direct identification with less original specimens, and achieve the effects of shortening the measurement time, simplifying the detection steps, and simplifying the detection process.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
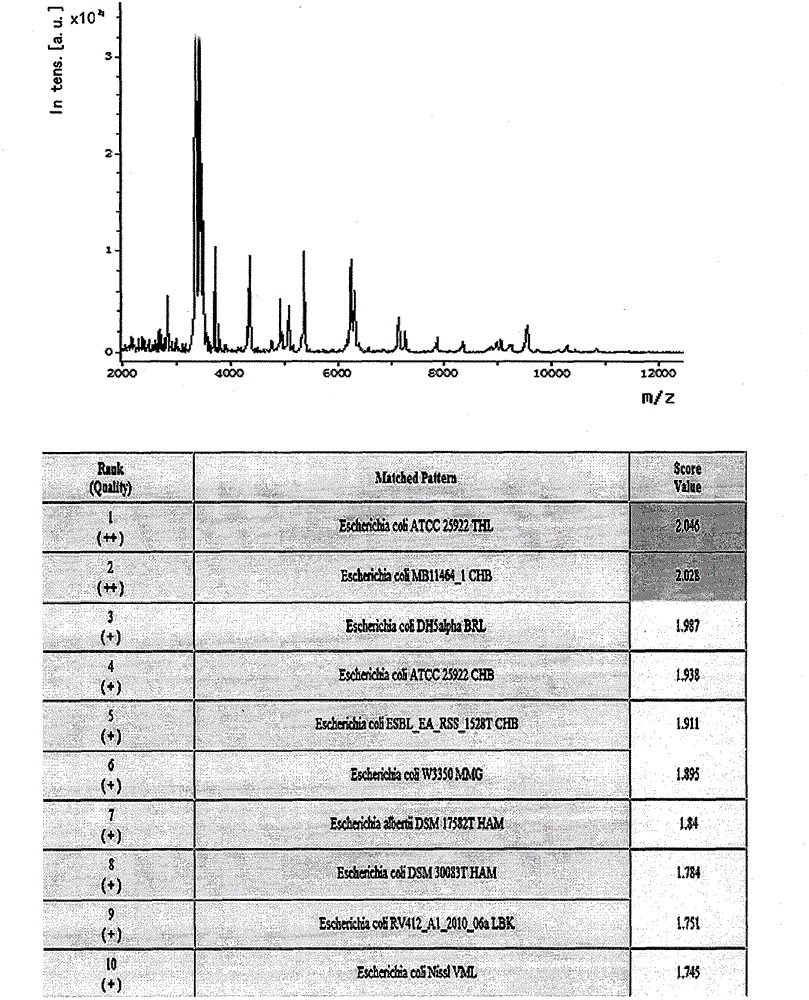
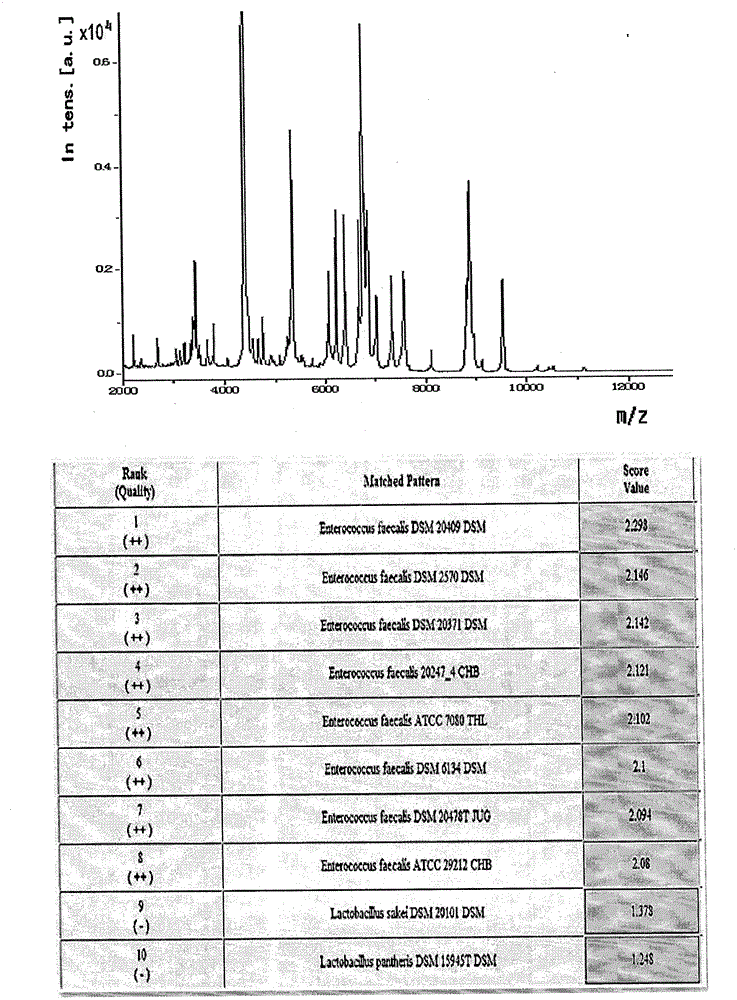
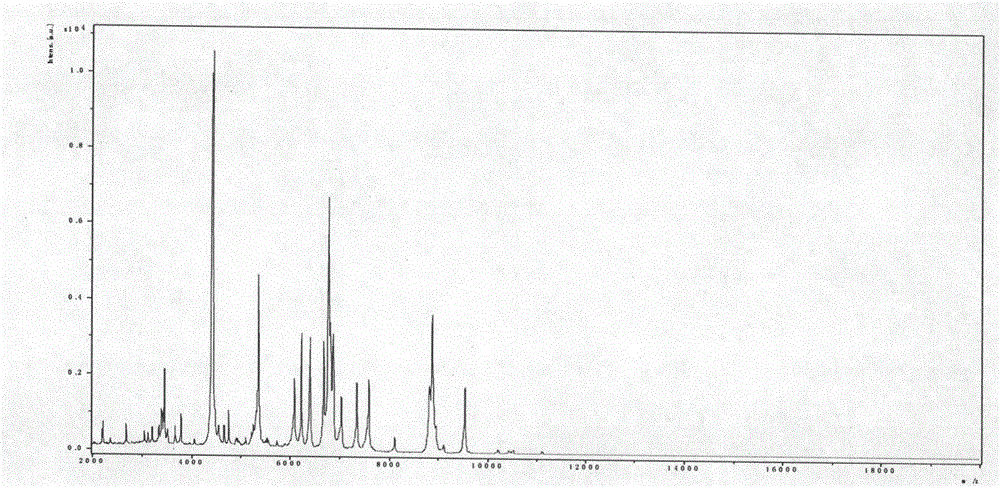
[0012] Example 1 MALDI-TOF MS detection sample pretreatment
[0013] Clinically, 1456 samples of clean midstream urine from patients with suspected urinary tract infection were collected and submitted for examination immediately. Take 1ml of urine sample at 2000×g, centrifuge for 30 seconds, and take the supernatant. Centrifuge at 15500rpm for 5 minutes to collect the precipitate. Wash once with sterile water, dry, and set aside. Add 5 μl formic acid (70%) and 5 μl acetonitrile for protein dissolution, mix well, take 1 μl spot target, add 1 μl matrix (α-cyano-4 hydroxycinnamic acid saturated solution, containing 50% acetonitrile and 2.5% trifluoroacetic acid), and tested on the machine after drying.
Embodiment 2
[0014] Embodiment 2 MALDI-TOF MS analysis
[0015] The acquisition of bacterial mass spectrometry requires autoflex III MALDI-TOF / TOF MS with a laser intensity of 200Hz and a detection range of 2-20kDa. Ion formation requires a 337nm nitrogen laser with a resolution delay time of 40ns. The detection of each sample requires 500 laser points to be collected and analyzed (10×50 laser irradiation points originate from different positions of the loading target). A mixture of protein standard 1 and peptide standard 2 is required for calibration before determination. The mass spectrogram is formed after the sample is measured, and analyzed by MALDI BioTyper software 3.0 and compared with the BioTyper bacterial database. The score range is 0-3, and the measurement score > 1.9 indicates that the species is identified; 1.7 < score < 1.9 indicates the identification to the genus; if the score is <1.7, the identification result is not credible.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com