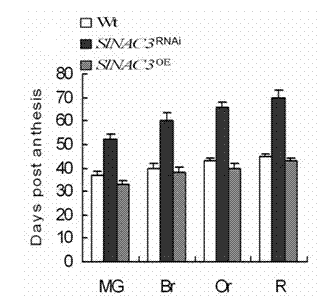
Tomato fruit ripening gene SINAC3 and application thereof
A fruit and mature technology, applied in the field of molecular biology and genetic engineering, can solve the problems of economic loss, easy softening and rot, etc., and achieve the effects of improving management level, short breeding cycle and cost saving
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1: SlNAC3 Cloning of full-length genes and 3'-specific fragments
[0024] The pericarp of tomato Ailsa Craig red ripe fruit was ground into powder with liquid nitrogen, then transferred to a centrifuge tube, and the total RNA was extracted by the guanidine isothiocyanate method. Reverse transcriptase M-MLV (Tiangen) was used to synthesize the first strand of cDNA using total RNA as a template. The reaction system and operation process were as follows: take 5 μg total RNA, add 50 ng oligo(dT) 15, 2 μL dNTP (2.5 mM each), add ddH at the end 2 O (no RNase) to a reaction volume of 13.5 μL; after mixing, heat denaturation at 70°C for 5 minutes, then quickly cool on ice for 5 minutes, then add 4 μL 5×First-stand buffer, 0.5 μL RNase inhibitor (200 U ), 1 μL reverse transcriptase (200U), 1 μL dimercaptothreitol (0.1M), mix well and centrifuge briefly, incubate at 42°C for 1.5h, take it out and heat at 95°C for 5min to terminate the reaction. After the first strand ...
Embodiment 2
[0028] Embodiment 2: plant expression vector construction
[0029] Construction of overexpression vectors: The cloning vector obtained in Example 1 and the plasmids of the plant expression vector pBI121 were extracted by alkaline lysis, and 1 μL was used for agarose gel electrophoresis to detect the integrity and concentration of the extracted plasmids. use Bam H I (Fermentas) and Xhol I (Fermentas) carried out double enzyme digestion on plasmids pMD-18T-SlNAC3 and pBI121 respectively (100 μL system). The reaction system and operation process were as follows: take 10 μL pMD-18T-SlNAC3 or pBI121 plasmids, add 10 μL 10×Tango buffer, 2 μL XbaI, 2 μL SalI, 76 μL ddH2O, mix well, centrifuge for a short time, and place at 37°C for overnight reaction. Spot all digested products on agarose gel for electrophoresis, and then SlNAC3 The fragment and the pBI121 large fragment were gel-recovered separately, and the whole process was performed using UNIQ-10 column DNA gel recovery ki...
Embodiment 3
[0034] Example 3: Genetic transformation of tomato
[0035] The genetic transformation system adopted is the genetic transformation method mediated by Agrobacterium, the Agrobacterium strain is C58, and the transformation recipient material is the wild tomato species Ailsa Craig. Soak plump tomato seeds in sodium hypochlorite solution (available chlorine content: 2%) for 15 min, then wash the seeds with distilled water and sow them on 1 / 2 MS medium, place at 25°C, and the photoperiod is 16 h light / 8 h Cultivate in the dark, and cut out the cotyledons when the sterile seedlings grow to 7-9 days old, and culture them in the dark on 1 / 2 MS medium for one day. Agrobacteria with a resuspended concentration of OD600≈0.5 were used to infect the leaves for 5 min, and the excess bacterial liquid was blotted dry with sterilized filter paper, and then re-placed on the pre-medium covered with filter paper, cultured in the dark for 2 days, and then transferred to the screening medium ( Ba...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com