SLAF-seq-based developed elytrigia elongata 1E chromosome specific molecular markers and application thereof
A long-spiked wheatgrass, specific marker technology, applied in application, plant genetic improvement, recombinant DNA technology and other directions, can solve problems such as no research on long-spiked wheatgrass, and achieve short cycle, good repeatability, and stable amplification. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Embodiment 1 Experimental material
[0046] Common wheat China spring (CS, 2n=6x=42), diploid Th. elongatum (Th. elongatum, EE, 2n=2x=14), China spring-Thn. elongatum addition line (DA lines), China Spring-Elongatum replacement lines (DS lines), Langdon (LD), Langdon-Elongatum diploid (8801), tetraploid Elongatum (Th.elongatum, 2n=4x=28) , Decaploid Elongated wheatgrass (Th.ponticum, PI179162, PI204383, 2n=10x=70), the above materials were donated by Dr.George Fedak of the Canadian Department of Agriculture. The original document of the materials is "Disomic and ditelosomic additions of diploidAgropyron elongatum chromosomes to Triticum aestivum", etc.; Yangmai 10 (Y10, formerly known as 93-111, bred by the Institute of Agricultural Sciences in Lixiahe, Jiangsu and Nanjing Agricultural University), Yangmai 14 (Y14, namely Yang 0-139, Jiangsu Li Yangmai 158 and Yangmai 6 used by Xiahe Agricultural Science Institute), Yangmai 16 (Y16, that is, Yang 0-126 is a hybrid of Y...
Embodiment 2
[0047] The extraction of embodiment 2 genome DNA
[0048] The test material grows to the stage of two leaves and one heart, and the genomic DNA is extracted by the SDS method. The steps are: (1) Take the young leaves (about 0.1g), cut them into pieces and put them in a 2ml centrifuge tube, place them in liquid nitrogen to cool, Grind to powder with a grinding rod;
[0049] (2) Place the centrifuge tube at room temperature to cool slightly, add 700 μl of buffer A (buffer A formula: 29.2g NaCl, 100ml of 1M Tris-HCl, 18.6g EDTA-Na, 15g SDS, ddH 2 O (dilute to 1L and sterilize), mix gently, then bathe in water at 65°C for 20 minutes, and mix by inverting up and down every 5 minutes during this period;
[0050] (3) Take it out and cool it down to room temperature, add 350l each of phenol and chloroform, turn it upside down, mix well, and extract for 5 minutes;
[0051] (4) 12000rpm, centrifuge for 10min, draw the supernatant into a new centrifuge tube;
[0052] (5) Add about 750...
Embodiment 3
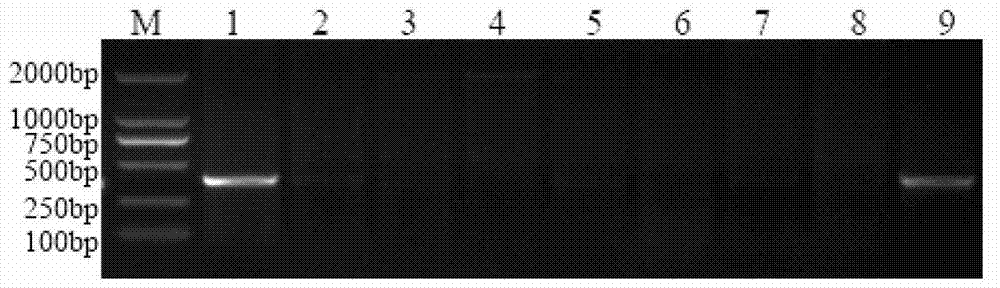
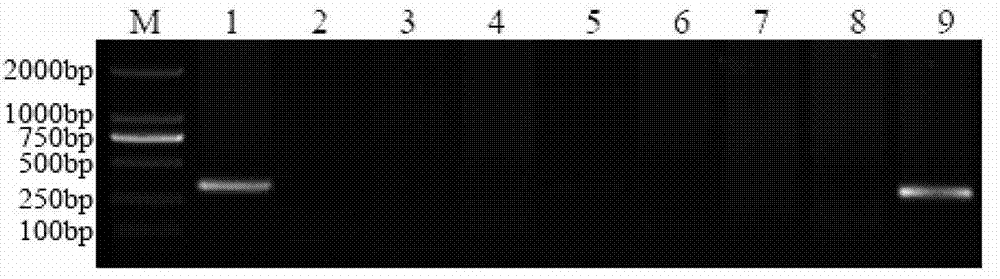
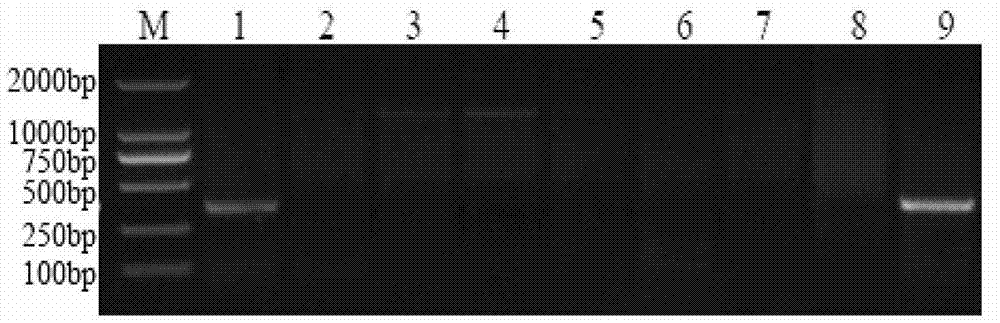
[0057] Example 3 Obtaining a Specific Fragment of Chromosome 1E of Echinopsis elongatum Based on SLAF-seq Technology
[0058] (1) Simplified protocol design: use enzyme digestion prediction software to analyze the reference genome system, design a marker development protocol based on the GC content of the genome, repeat sequence conditions, and gene characteristics, and determine the enzyme digestion protocol, gel cutting range, and sequencing volume etc. to ensure the density and uniformity of its molecular marker development so as to ensure that the expected experimental purpose is achieved.
[0059] (2) Enzyme digestion of genomic DNA: Select the target fragment by enzyme digestion to meet the expected design index requirements for the number of tags. The enzyme digestion system is 500ng of genomic DNA (genomic DNA of Chinese spring, Ethiopyrum elongatum, and Chinese spring-Ethiopyrum elongatum 1E addition line), 1 μl of NEB buffer4 (buffer 4 from New England Biolabs), 0.12...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com