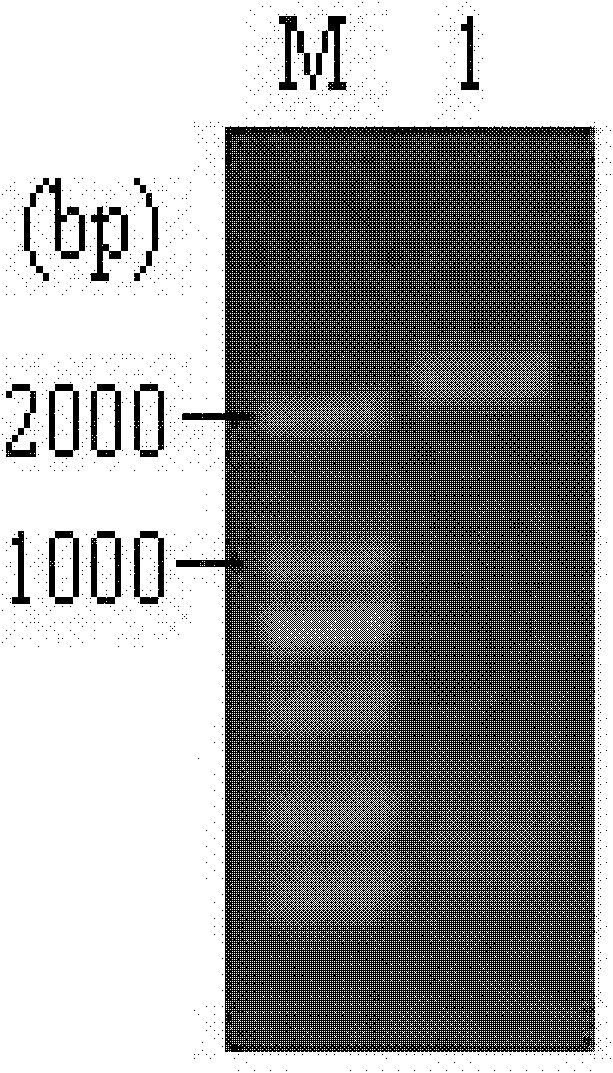Alpha-galactosidase, and coding gene and application thereof
A galactosidase and gene technology, which is applied to α-galactosidase and its encoding gene and application fields, can solve the problems of few research reports on α-galactosidase, and achieve great application potential and good thermal stability. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Embodiment 1, the discovery of α-galactosidase and its coding gene
[0064] Table 2 Primers used for cloning of α-galactosidase coding gene
[0065] Primer
5′→3′
Length (bp)
galDF
ATHAARTGGGAYATGAA
17
galDR
TCNGGNCKNGTRTTRTC
17
gal5′GSP
GCCACTGGAGCAGGTTTCAATC
22
gal5'NGSP
CGCTTATAGTGGTGCATGGTGA
22
gal3′GSP
GAACAGACCGTTTGCCGAAGTC
22
gal3'NGSP
TGCTGATTGAAACCTGCTCCAG
22
[0066] Note: In the above degenerate primers galDF and galDR, H=A / C / T, R=A / G, Y=C / T, N=A / T / C / G, K=G / T.
[0067] A pair of primers were designed according to the existing homologous sequences and conserved regions, consisting of galDF and galDR (see Table 2). The genomic DNA of Rhizomucor miehei (Rhizomucor miehei) CAU432 was used as a template, and the primer pair consisting of galDF and galDR was used for PCR amplification. PCR amplification conditions: 95°C for 5min; 95°C for...
Embodiment 2
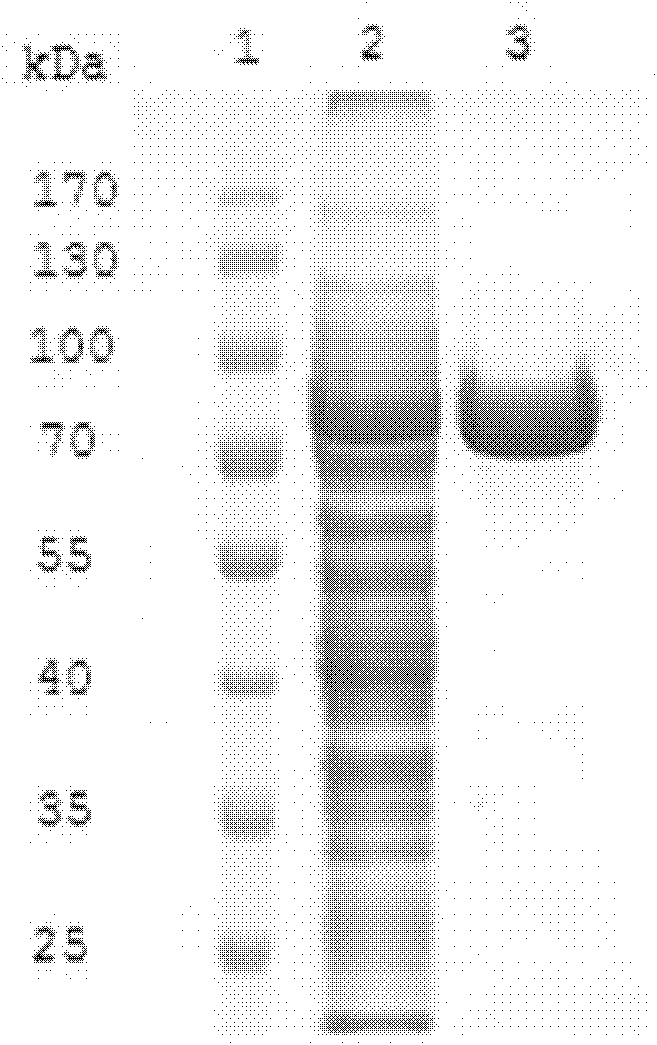
[0071] Embodiment 2, expression and purification of recombinant α-galactosidase gene
[0072] 1. Construction of recombinant expression vector
[0073] 1. Extract the total RNA of Rhizomucor miehei CAU432 and reverse transcribe it into cDNA.
[0074] 2. Using the cDNA in step 1 as a template, carry out PCR amplification with a primer pair consisting of RmgalAF (38bp) and RmgalAR (45bp), and reclaim the PCR amplification product. The agarose gel electrophoresis of the PCR amplification product is shown in figure 1 (M is DL2000marker, 1 is PCR amplification product).
[0075] RmgalAF: 5′-GGAATTC CATATG CTGGACACTGGCATCCACAAGCACC-3′;
[0076] RmgalAR: 5′-ATAAGAAT GCGGCCGC TTTCTTTTGTACCCAAAACAACAGCGCTTC-3'.
[0077] 3. The PCR amplified product was double-digested with restriction endonucleases Nde I and Not I, and the digested product was recovered.
[0078] 4. Digest the pET-30a(+) vector with restriction endonucleases Nde I and Not I to recover the vector backbone (about...
Embodiment 3
[0106] Embodiment 3, Escherichia coli produces the character of recombinant α-galactosidase
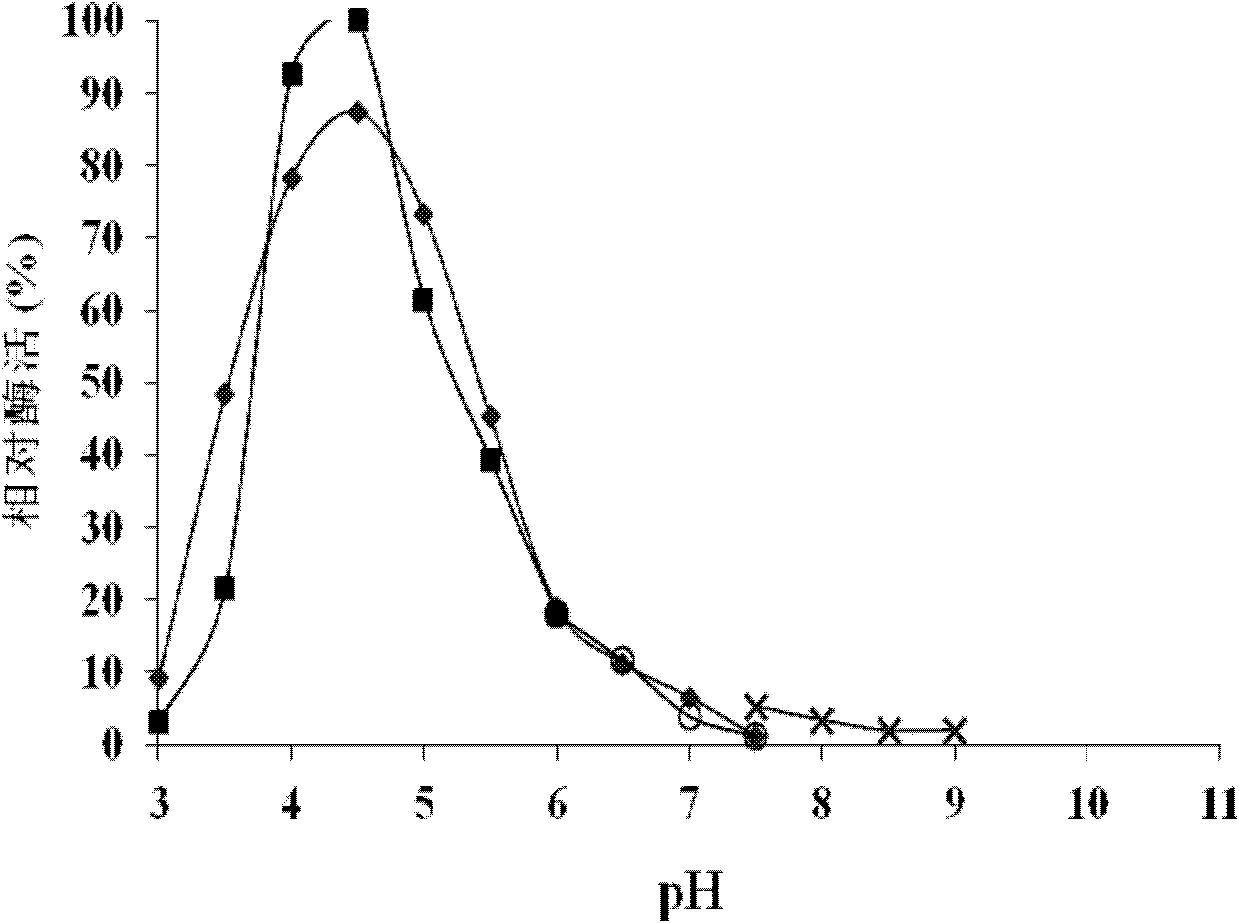
[0107] 1. Determination of optimum pH
[0108] The RmgalA protein solution prepared in Step 3 of Example 2 was used as the solution to be tested.
[0109] Enzyme assay method is the same as the 2 of the step five of embodiment 2, and difference only is to adopt following several damping solutions respectively (concentration is 50mM): citric acid-phosphate buffer (pH 3.0-7.5), citric acid buffer (pH 3.0-6.0, phosphate buffer (pH 6.0-7.5), Tris-Cl buffer (pH 7.5-9.0), CHES buffer (pH 8.0-10.0), Glycine / NaOH buffer (pH 9.0-11.0).
[0110] Citric acid-phosphate buffer solution (indicated by "◆"): prepare 50mM disodium hydrogen phosphate and 50mM citric acid respectively, and mix the two according to different ratios to adjust the pH to pH 3.0-7.5.
[0111] Citric acid buffer (indicated by "■"): Prepare 50mM citric acid and sodium citrate solutions respectively, mix the two in different ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com