Type two peptidoglycan recognition protein and preparation method and application thereof
A technology for identifying proteins and peptidoglycan, which is applied in the fields of peptide/protein components, biochemical equipment and methods, medical preparations containing active ingredients, etc. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] The type II peptidoglycan recognition protein is shown in the amino acid sequence of SEQ ID No.1 in the sequence table. The sequence listing SEQ ID No.1 is:
[0021] MTSFGVSIFVLGLSCFFTVYSRPAGVHLRNMDSFIRAVQQVEDSNPGISPLALVRALRRTAGHD
[0022] DAMTIHFLGASYNLSDAEVLETAILNASSFSFFDKAIHHIVTDYGEERGVVLTPDGTTVALAPL
[0023] LLG I ELGLKAKMEGTPAVGLFSLTLRTLGLSFLSLQDFPPSYRLGPNGCWDNVDYPKMFKLSQ
[0024] AATLATDAVINGGMDGAILGT
[0025] DLSNLSAHEPPQALSDILKGYYSFILHEGQGLDALTKHVSPRR
[0026] REISRAILEPLDLHSQVMETLALVWKLEKTEWIAFDTGLGKAVKDGLERFVHKYWDCPQIIPRC
[0027] QWGAKAHRGTPPLPLSLPLRFLYVHHTYEPSSPCLSFPTCSRDMRSMQRYHQEDRGWSDIGYSFV
[0028] VGSDGYVYEGRGWNQLGRHTRGHNDVGYGVSIIGNYTATLPSRHAMDLLLRHRLVRCAVDGGGLV
[0029] ANFTIHGHRQVVNYTSCPGDAFFSEIRTWEHFRD
[0030] (a) Sequence features:
[0031] ●Length: 482
[0032] ●Type: amino acid sequence
[0033] ●Chain type: single chain
[0034] ●Topological structure: linear
[0035] (b) Molecule type: protein
[0036] (c) Assumption: No
[0037] ...
Embodiment 2
[0041] The preparation method of type II peptidoglycan recognition protein:
[0042] 1) Construction of plasmid pPGRP2:
[0043] Using American redfish cDNA as a template, PCR amplification was performed with primers PGRPF1 and PGRPR1. The PCR conditions are: 94°C for 60s to pre-denature template DNA, then 94°C for 40s, 55°C for 60s, 72°C for 60s, after 5 cycles, change to 94°C for 40s, 61°C for 60s, 72°C for 60s, and after 25 cycles, repeat at 72°C. ℃ extension reaction 7-10min. After the PCR product was purified, it was ligated with the carrier pBS-T (purchased from "Tiangen Biochemical Technology (Beijing) Co., Ltd.") at room temperature for 2-8 hours, and the ligation mixture was transformed into E. ml Xgal and isopropyl-β-D-thiogalactoside (24ug / ml) were cultured on LB medium for 18-24 hours, and the transformants were screened to extract a plasmid, which was plasmid pBSPGRP1.
[0044] The above-mentioned plasmid pBSPGRP1 and plasmid pET259 (see Zheng, W.J., Hu, Y.H., ...
Embodiment 3
[0051] Bactericidal effects of type II peptidoglycan recognition proteins
[0052] 1) Bacterial plate preparation. Streptococcus iniae, Staphylococcus aureus and Bacillus subtilis (all preserved in CGMCC, numbered CGMCC No.1984, 1.363, 1.460 respectively) were cultured in LB medium to OD 600 0.5, the bacterial solution was mixed with LB containing 0.8% (mass percentage) agar; the mixed solution was poured into a petri dish, and it was a bacterial plate after solidification.
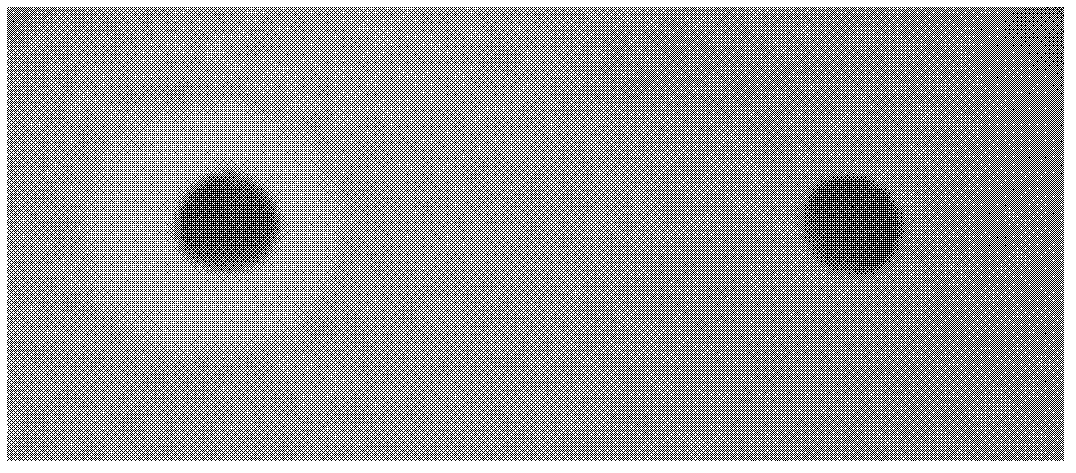
[0053] 2) Detection of the bactericidal effect of the type II peptidoglycan recognition protein. Place the piece of sterile filter paper on the bacterial plate of 1) above. The peptidoglycan recognition protein prepared in Example 2 was diluted to 5uM in PBSZ, and 10ul of the peptidoglycan recognition protein or PBSZ (negative control) was added to the filter paper of the bacterial plate. After the plate was incubated at 28°C for 12-14 hours, it was observed that an obvious inhibition zone was formed a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com