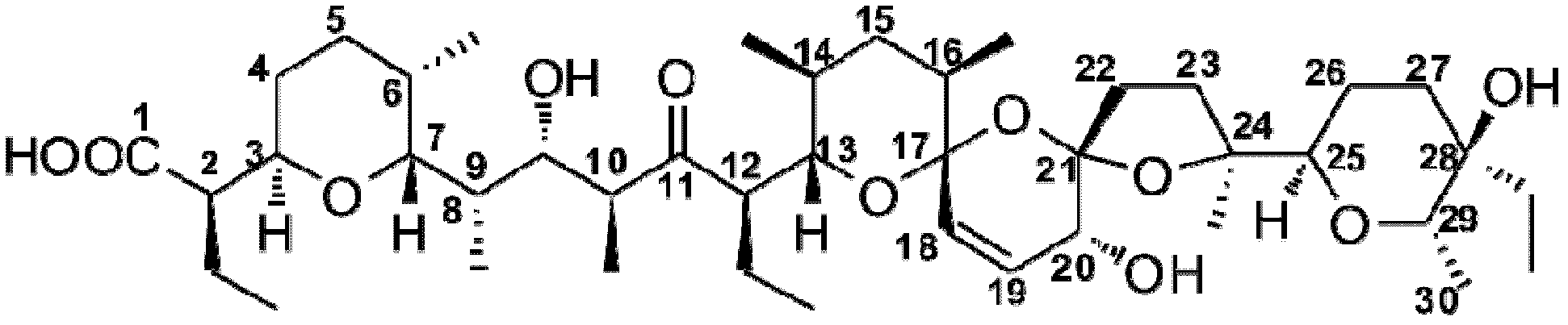
Biosynthetic gene cluster of salinomycin and application thereof
A biosynthesis and gene cluster technology, applied in the field of microbial genetic resources and genetic engineering, can solve the problems of salinomycin biosynthesis pathway not clarified
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0124] The biosynthetic gene fragment of embodiment 1 cloning salinomycin
[0125] Design degenerate primers based on the conserved amino acid sequence of cyclooxygenase in the reported polyether biosynthetic gene cluster:
[0126] SEQ ID NO.32 (Epo-110-F-2): 5'-GTS ACC GTS RTS GAV CGN GA-3';
[0127] SEQ ID NO.33(Epo-1030-R-1): 5'-GCT CAT SCC RTG SCC GT-3'
[0128] Wherein, S=C / G, R=A / G, N=hypoxanthine.
[0129] The partial sequence of the cyclooxygenase gene was cloned from the total DNA of the salinomycin-producing strain Streptomyces albus XM211 to obtain a specific PCR product with the same size as the expected fragment (about 0.9kb), cloned into the pMD18-T vector, and analyzed by DNA sequencing It shows that it has high homology with known cyclooxygenase coding genes, and can be used as a probe to screen the constructed genome library.
Embodiment 2
[0130] Cloning and analysis of embodiment 2 salinomycin biosynthesis gene cluster
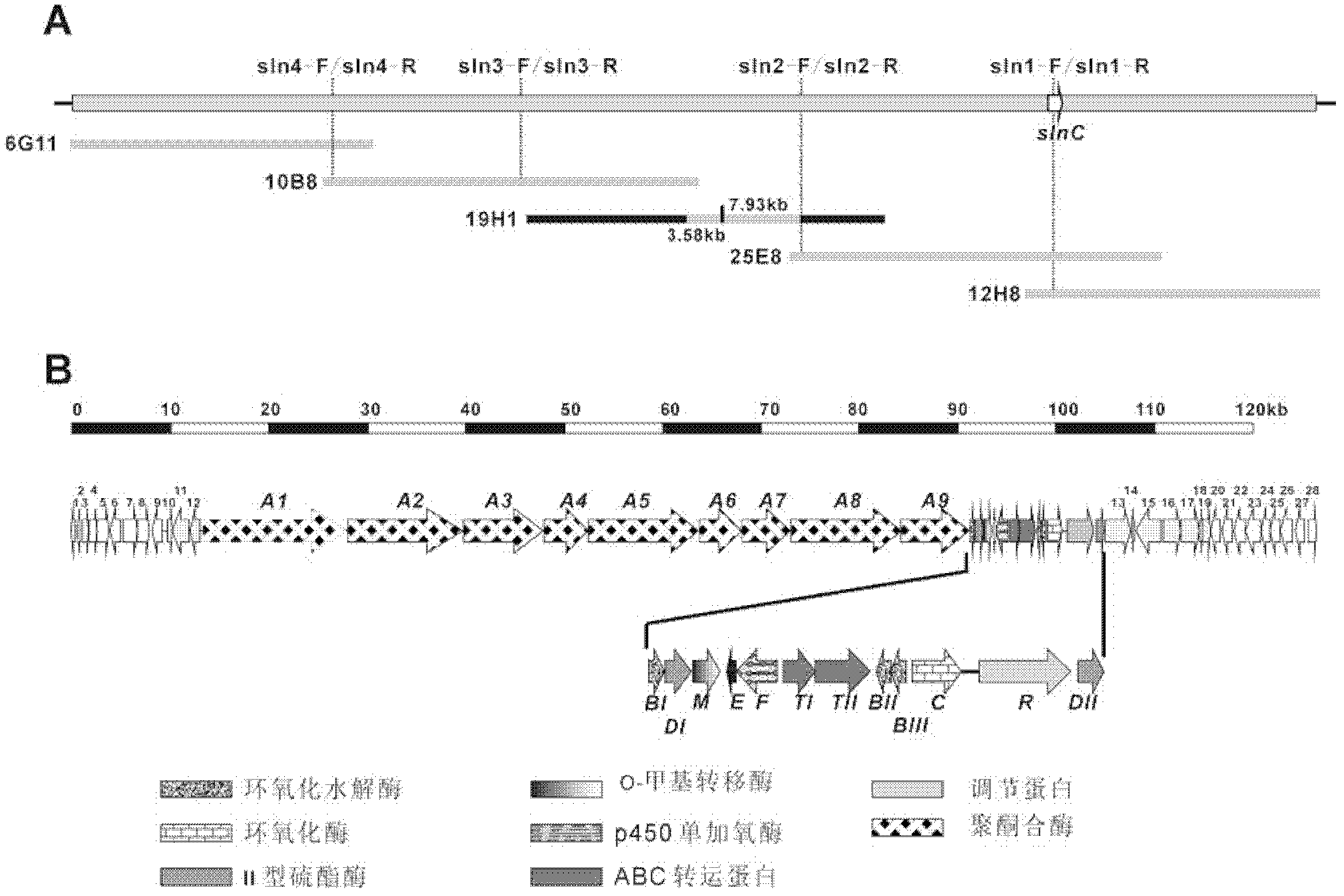
[0131] Using the cyclooxygenase gene fragment obtained in Example 1 as the probe sln1-F / R, 12 cosmids were isolated from approximately 3000 clones in the genome library of Streptomyces albusXM211. One of the cosmids, 12H8, was selected for subcloning and sequencing, and multiple sequence information obtained were in line with expectations, and its full-length sequencing was performed.
[0132] On this basis, 3.58 kb and 7.93 kb sequences of four cosmids 12H8, 25E8, 10B8, 6G11 and 19H1 were obtained by chromosome walking and sequencing, covering about 128 kb of the chromosome ( figure 2 A).
[0133] Bioinformatics analysis of the sequence contained 49 open reading frames ( figure 2 B), which contains all polyketide synthase genes (PKS), modification genes (modification), partial regulation genes (regulation) and transport-related genes (transporter) necessary for the synthesis of salinomycin...
Embodiment 3
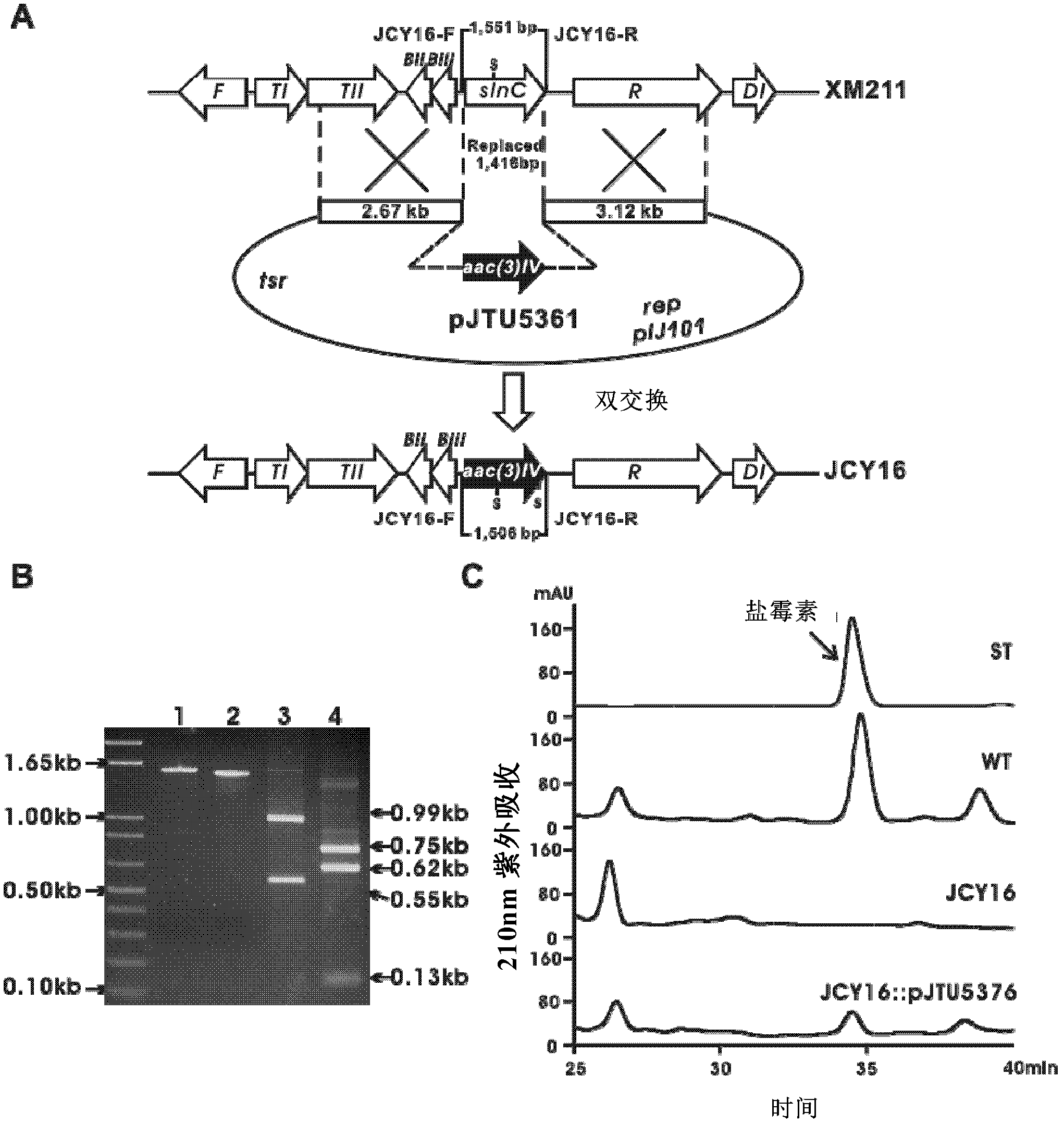
[0140] Example 3 Interruption of Cyclooxygenase Gene sln C
[0141] In order to verify that the cloned region is related to the biosynthesis of salinomycin, the 1.416bp region (inside the cyclooxygenase gene) in the first Fosmid 12H8 screened by degenerate primers was replaced.
[0142] Since the aac(3)IV (Apramycin resistance gene) gene is inserted into the slnC gene, a 1.55kb PCR amplification fragment was amplified from the genomic DNA of the Streptomyces albus XM21 wild-type strain using primers JCY16-F / R , and the mutant strain JCY16 amplified a 1.50kb fragment, which was verified by digestion with the SacI restriction enzyme site inside the amplified fragment, and the results obtained were also in line with expectations, that is, the wild-type strain obtained two fragments of 0.55kb and 0.99kb Fragment, while the mutant strain got 0.62kb, 0.75kb and 0.13kb three fragments ( image 3 B).
[0143] The results of HPLC detection of fermentation products showed that the Str...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com