Real time pcr detection of single nucleotide polymorphisms
A real-time detection and polymorphism technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, instruments, etc., can solve the problems of poor sensitivity, cost and time consumption, unsuitable for high-throughput applications, and unsatisfactory real-time detection. , to achieve the effect of user-friendly and reliable, fast detection method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
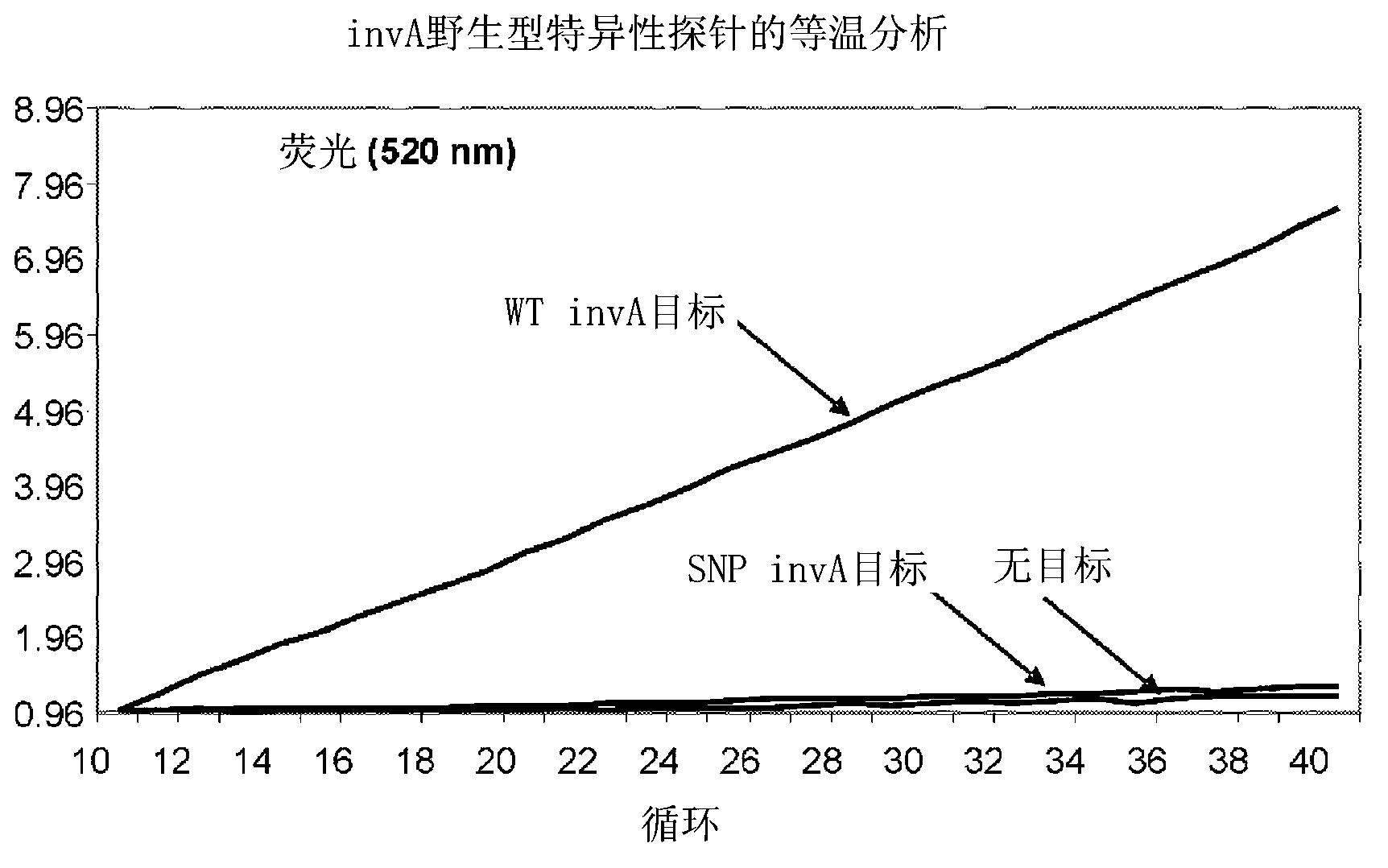
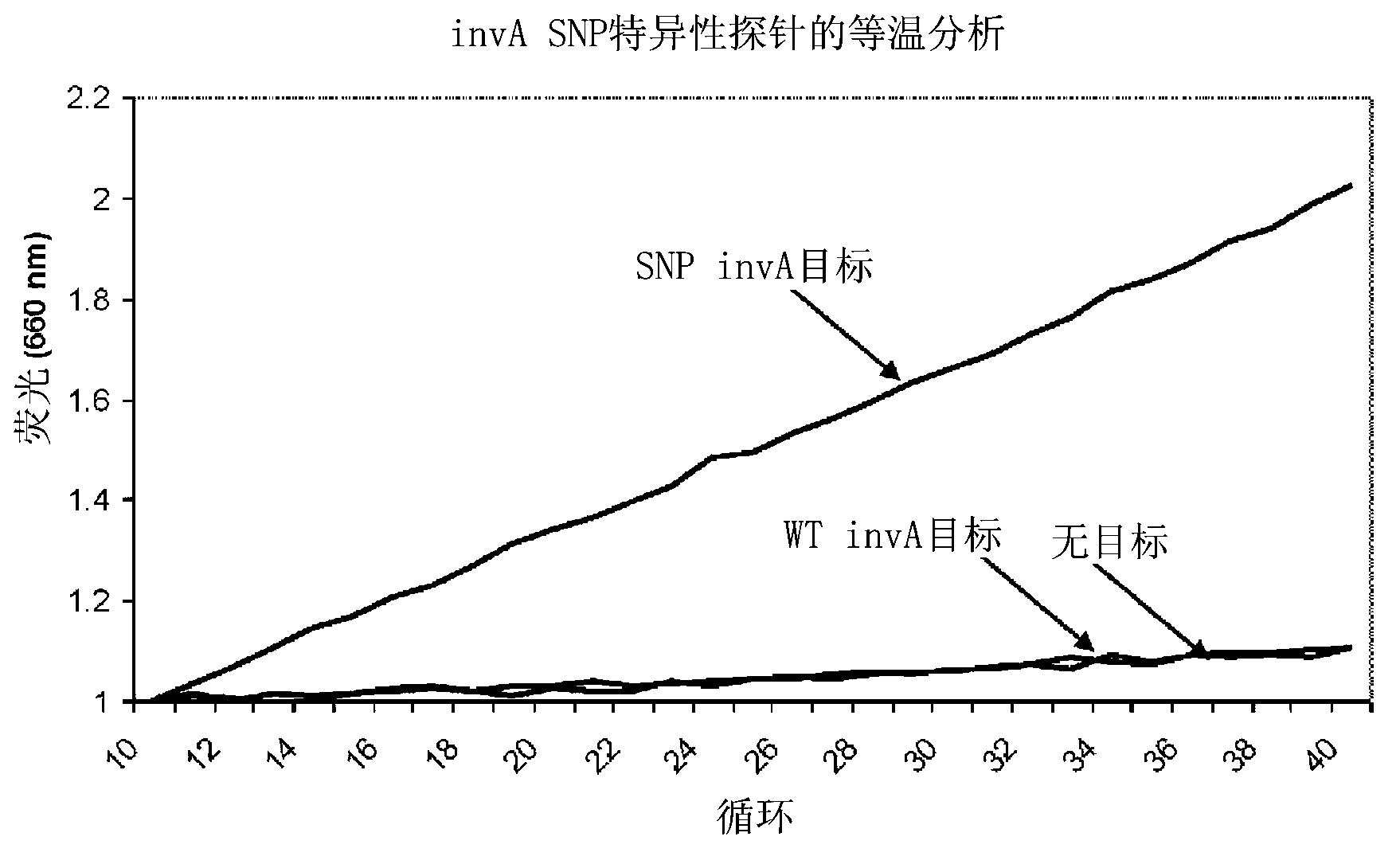
[0189] Example 1: Isothermal detection of synthetic SNPs in the Salmonella invA gene
[0190] Construction of artificial single nucleotide polymorphisms (SNPs) in the invA gene (SEQ ID NO: 33) of Salmonella to test CataCleave TM The ability of a probe to distinguish single nucleotide sequence differences in a target DNA sequence. A single nucleotide change produced a T to G transversion at position 116 of the SalmonellainvA coding sequence (SEQ ID NO: 33). Design of two similar 19-nucleotide CataCleave TM Probes, wherein each probe is doubly labeled to generate FRET pairs such that the probes will base pair across the region of invA comprising the SNP nucleotide. The wild-type specific probe inv-CC probe 2 (SEQ ID NO: 1) is completely complementary to the wild-type sequence of invA, and the SNP-specific probe inv-CC probe 2-2C (SEQ ID NO: 2) is completely complementary to the invA The mutant form is fully complementary. Design probes such that CataCleave TM The second of ...
example 2
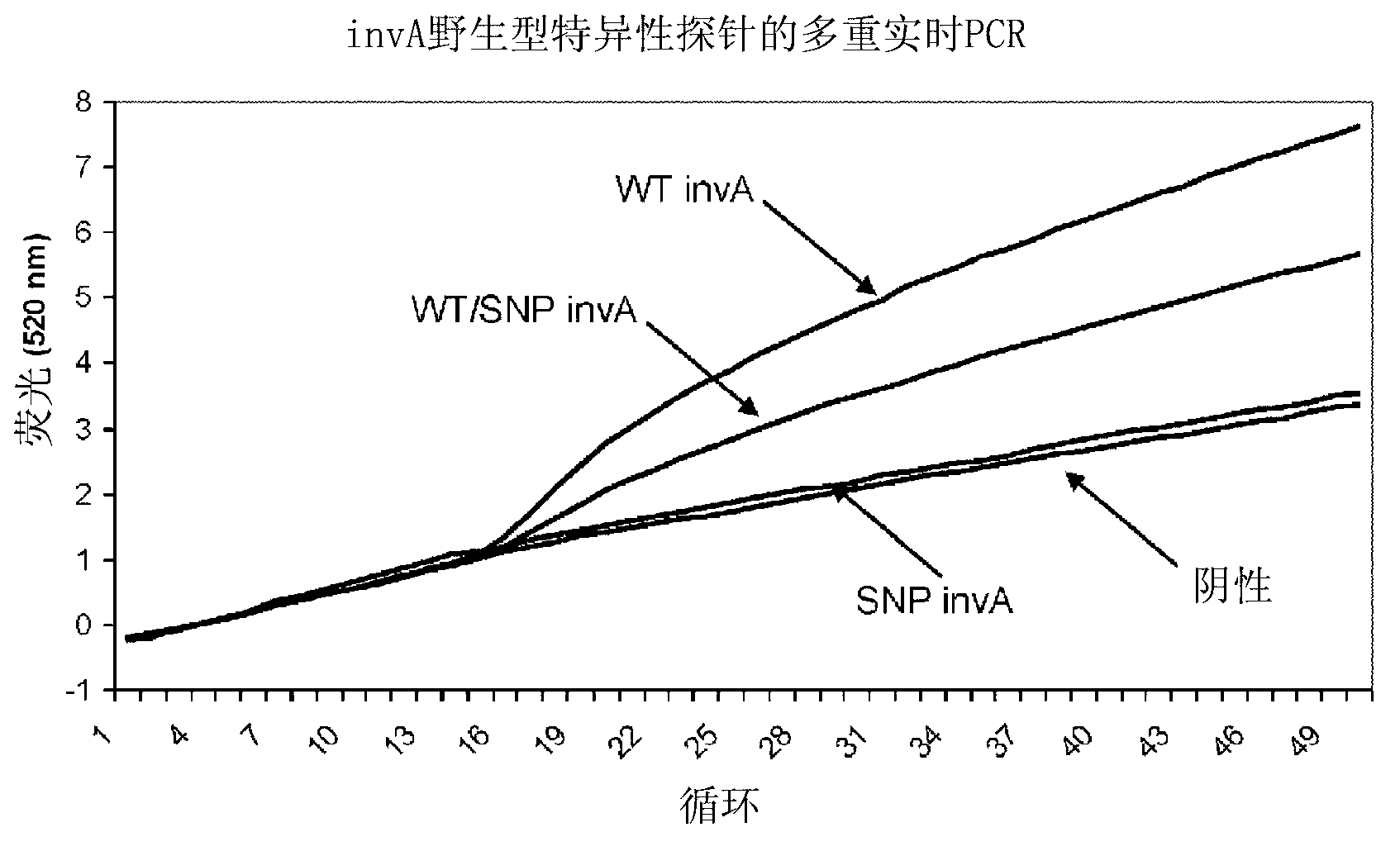
[0192] Example 2: Real-time PCR detection of synthetic SNPs in the Salmonella invA gene
[0193] Plasmid DNA containing a 267 nucleotide invA sequence containing wild-type or mutated bases (described above) was synthesized. 40 pg of wild-type plasmid, mutant plasmid or a mixture of both plasmids was used as template for multiplex real-time PCR reactions containing differentially labeled probes complementary to wild-type or mutant sequences. The final concentration of each component in the reaction was as follows: 800 nM forward primer Salmonella-F1 (SEQ ID NO: 5), 800 nM reverse primer sal-invR2 (SEQ ID NO: 6), 200 nM wild-type specific probe Needle inv-CC probe 2 (SEQ ID NO: 1), 200 nM SNP-specific probe inv-CC probe 2-2C (SEQ ID NO: 2), 80 uM each dNTP, 10 mM Tris acetic acid (pH 8.6) , 50 mM Potassium Acetate, 2.5 mM Magnesium Acetate, 1 mM DTT, 2.5u Platinum Taq DNA Polymerase (Life Technologies) and 2.5u Hybridase Thermostable RNase HI (Epicentre). The PCR reaction was ...
example 3
[0194] Example 3: Isothermal detection of the A1 and A2 alleles of the bovine b casein gene
[0195] Two completely natural variants or forms of b casein exist in the milk of cows, called A2 and A1b casein. The difference between A1 and A2b caseins is a single amino acid at position 67. In the A1 variant, the base is T, and in the A2 variant, the base is G. The A1 variant b casein in milk is unique among all mammalian b caseins in having a histidine at this position. Milk of other species containing beta-casein may be considered A2 category due to having proline at the equivalent position in their beta-casein chain. Buffalo, yak, goat, and human breast milk all contain the A2-form of beta-casein.
[0196] In this example, two similar 19-nucleotide CataCleaves were designed TM Probes, wherein each probe is doubly labeled to generate FRET pairs such that they will base pair across the position of the A1 / A2 SNP nucleotide in the bovine b casein gene. A1-CC probe 2-RC (SEQ ID...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com