Extreme halophilic archaea engineering bacteria for producing bioplastics PHBV by effectively utilizing carbon source
An extremely halophilic archaea, sequence technology, applied in the directions of microorganism-based methods, microorganisms, genetic engineering, etc., can solve the problems of increasing the viscosity of the culture medium, increasing the amount of foaming, and wasting carbon sources, and solve the problem of the culture medium becoming viscous. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Example 1. Determination of key gene clusters for exopolysaccharide synthesis and acquisition of genes
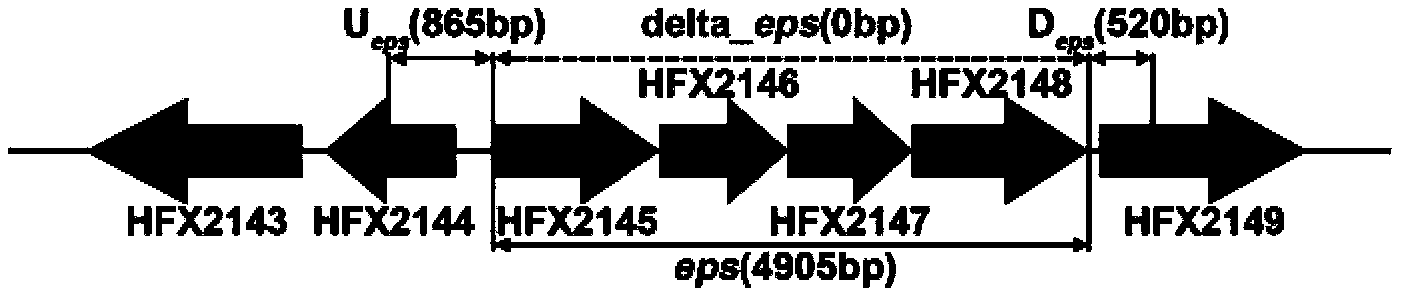
[0059] The complete set of glycosyltransferases is summarized based on the whole genome sequence and annotations of Halobacterium mediterranei. After analysis, the gene HFX2145-2148 was considered as a possible key gene cluster for exopolysaccharide synthesis (eps). These four genes may share a promoter. Among them, HFX2145 is involved in the biosynthesis of the precursor substance UDP-N-acetylglucosamine, HFX2146 and HFX2147 are involved in the sequential transfer of sugar groups to the plasma membrane carrier dolicol phosphate, and HFX2148 is involved in the trisaccharide monosaccharide body transmembrane transport. The structure of the four coding genes and the arrangement of the upstream and downstream genes are as follows: figure 1 shown.
[0060] A pair of primers epsF1 / epsR1 was designed for PCR amplification of the nucleotide sequence including the key gen...
Embodiment 2
[0067] Example 2. Identification of eps function of the key gene cluster for exopolysaccharide synthesis
[0068] Knock out and complement eps by genetic engineering technology, and verify that it is necessary for exopolysaccharide synthesis by genetic methods.
[0069] 1. Construction of uracil synthesis-deficient exopolysaccharide synthesis key gene cluster (eps) knockout strain
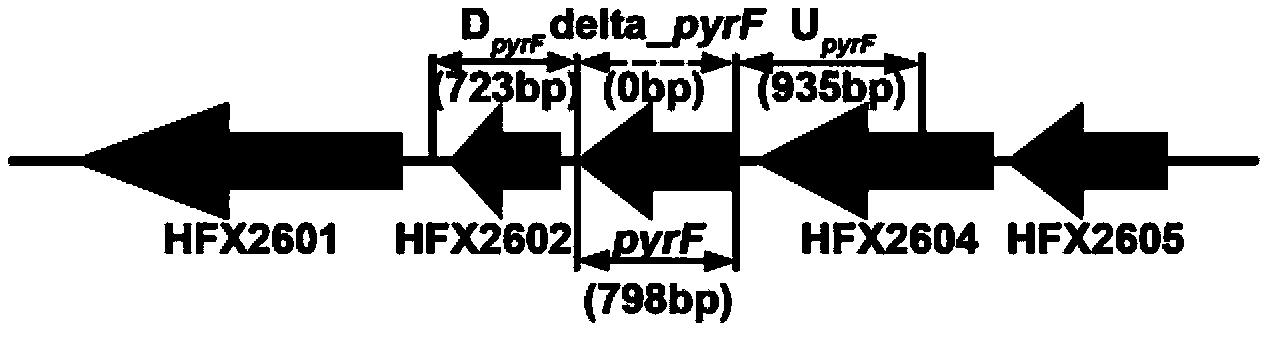
[0070] Using the genetic operating system based on the pyrF gene, the structural diagram of the pyrF gene and its upstream and downstream sequences is shown in figure 2 As shown, using Haloferax mediterranei ΔpyrF (Haloferax mediterranei ΔpyrF is the Mediterranean haloferax mediterranei CGMCC 1.2087 as the starting strain, the uracil synthesis-deficient strain obtained by knocking out the pyrF gene is convenient for the screening of gene knockout and complementation, and improves the efficiency of knockout. Knockout efficiency; Liu,H.,J.Han,et al.(2011)."Development of pyrF-based gene knockout sy...
Embodiment 3
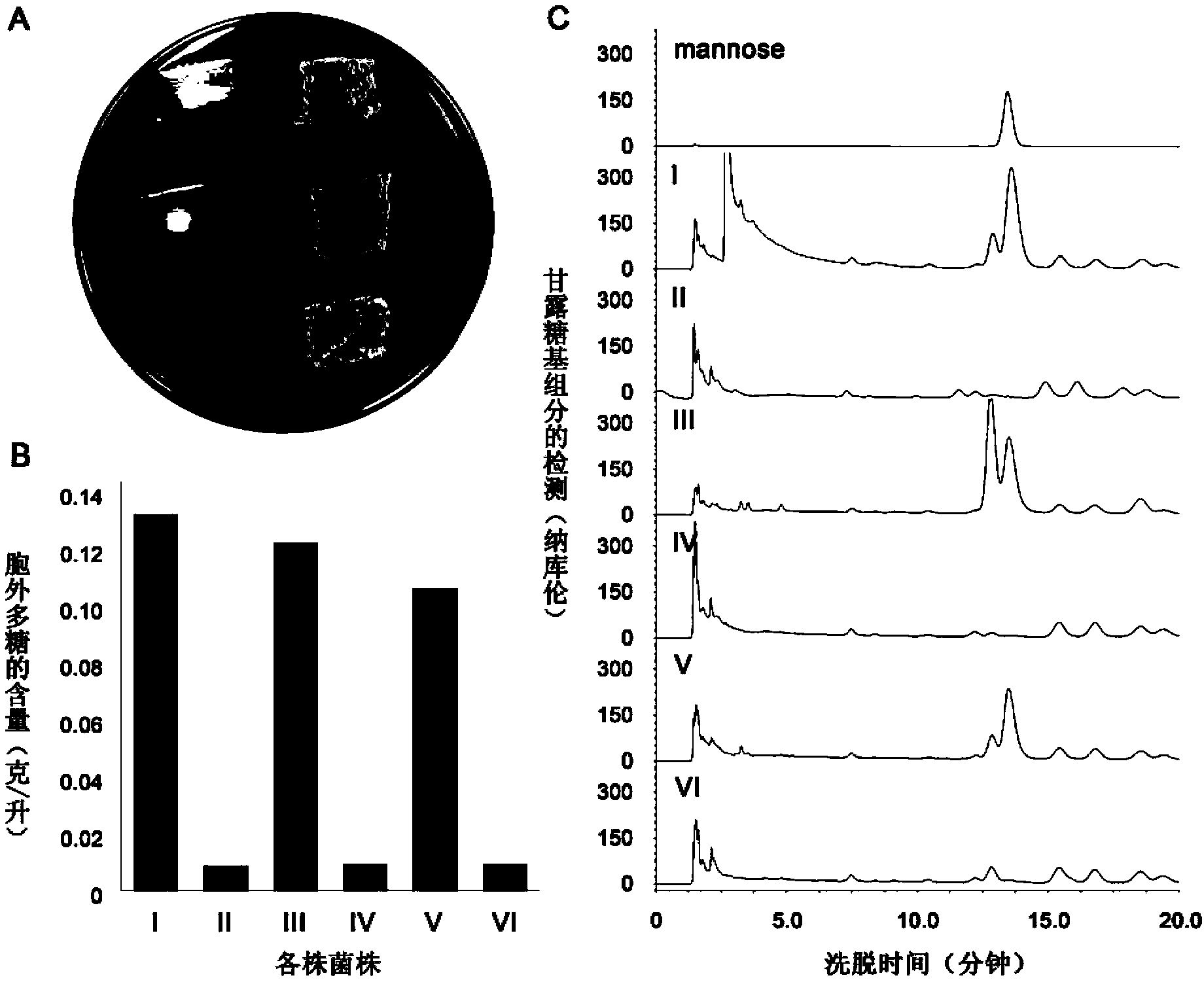
[0121] Example 3, Construction of eps knockout strain independent of exogenous addition of uracil growth
[0122] The eps knockout strain Haloferax mediterranei ΔpyrFΔeps, which is deficient in uracil synthesis, relies on exogenously added uracil. Even though it can efficiently use carbon sources to produce PHBV, it is difficult to be used as a strain for industrial production. Therefore, it is necessary to complement its missing gene pyrF in situ.
[0123] 1. In situ knock-in of deletion gene pyrF in strains deficient in uracil synthesis
[0124] Using the genetic operating system based on the pyrF gene, Haloferax mediterranei ΔpyrFΔeps was used as the starting strain, and the deleted gene pyrF was knocked in in situ to construct an eps deletion mutant that does not rely on exogenous addition of uracil. First construct the integrated plasmid pT-pyrFr for in situ knock-in of pyrF, and then undergo homologous recombination single crossover and homologous recombination double cr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com