Increased production of 2,3-butanediol in bacillus amyloliquefaciens by enhancing the regeneration rate of the coenzyme cycle
A recycling and butanediol technology, applied in the field of genetic engineering, can solve problems that do not conform to industrial safety production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0011] Embodiment 1: Amplification of target gene and construction of recombinant Bacillus amyloliquefaciens
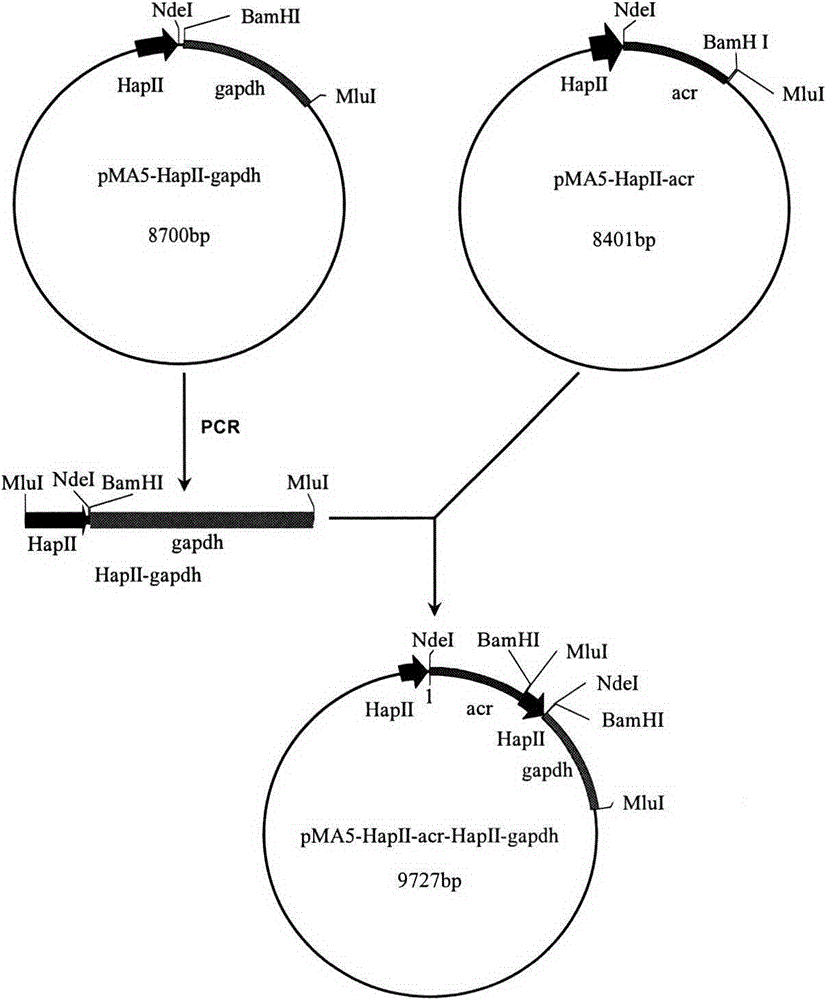
[0012] The schematic diagram of the construction of plasmid pMA5-HpaII-acr-HapII-gapdh is as follows figure 1 As shown, the specific process is as follows:
[0013] First, using the chromosomal DNA of the bacterial strain B10-127 as a template, using primers P1 and P2, a 901bp nucleotide sequence of the acr gene shown in SEQ ID NO: 1 was amplified by PCR technology, and the purified acr After the gene was digested by restriction endonucleases NdeI and BamHI, it was ligated with the plasmid pMA5-HapII which was also digested by the above two restriction endonucleases (the sequence of pMA5-HapII is shown in SED ID NO: 3), and the recombinant plasmid pMA5 was constructed -HapII-acr, verified by double enzyme digestion, indicating that the recombinant plasmid was constructed successfully. Subsequently, using primers P3 and P4 to obtain the gene gapdh whose nucleotide se...
Embodiment 3
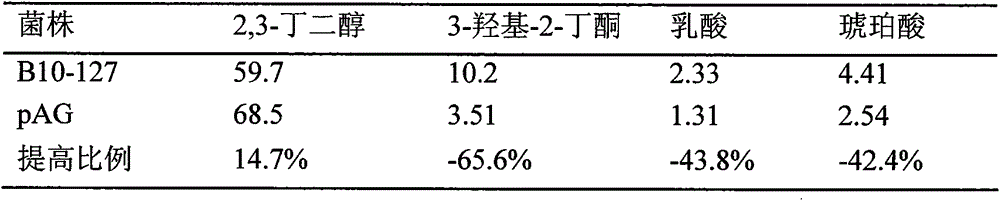
[0020] Embodiment 3: Fermentation performance verification of bacteria original bacteria and recombinant bacteria
[0021] (1) Seed cultivation
[0022] Pick a single colony from the activated plate and inoculate it into the seed medium. The seed culture temperature is 37°C, the shaker speed is 160r / min, and the culture time is about 12h. The composition of the seed medium: yeast extract 5g / L, tryptone 10g / min L, NaCl10g / L.
[0023] (2) Fermentation culture
[0024] The initial fermentation culture volume is 2.5L, and the components of the fermentation medium used are as follows:
[0025] Fermentation medium composition: glucose 160g / L, corn steep liquor 5g / L, urea 3g / L, sodium citrate 6g / L, K 2 HPO 4 4g / L, MgSO 4 0.2g / L; adjust the pH of the above fermentation medium to 6.5 with 5mol / L NaOH, and sterilize at 121°C for 30min.
[0026] Fermentation Fermentation conditions: inoculate the above-mentioned cultivated seed liquid into the fermentation medium according to the i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com