Application of G6PDH gene for improving steroid C11 alpha-hydroxylation ability of rhizopus nigricans and strain
A technology of rhizopus niger and hydroxylation, applied in the fields of application, genetic engineering, plant gene improvement, etc., can solve problems such as difficult conversion rate, achieve high application value, high conversion rate and conversion efficiency, and far-reaching theoretical significance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1: Related primer design
[0035] Primer F-g6pdh / R-g6pdh and primer F-hy g were designed according to the known Rhizopus oryzae G6DPH gene sequence (SEQ ID No.1) and E.coli hygromycin resistance gene sequence (SEQ ID No.2) / R-hyg (see Table 1), BamHI and ApaⅠ restriction sites were added to the two ends of primer F-g6pdh / R-g6pdh respectively, and primer F-hyg / R-hyg was used to identify Rhizopus niger transformants by PCR.
[0036] Table 1: Primers involved in the present invention
[0037] Primer name
Embodiment 2
[0038] Embodiment 2: the extraction of Rhizopus oryzae total RNA
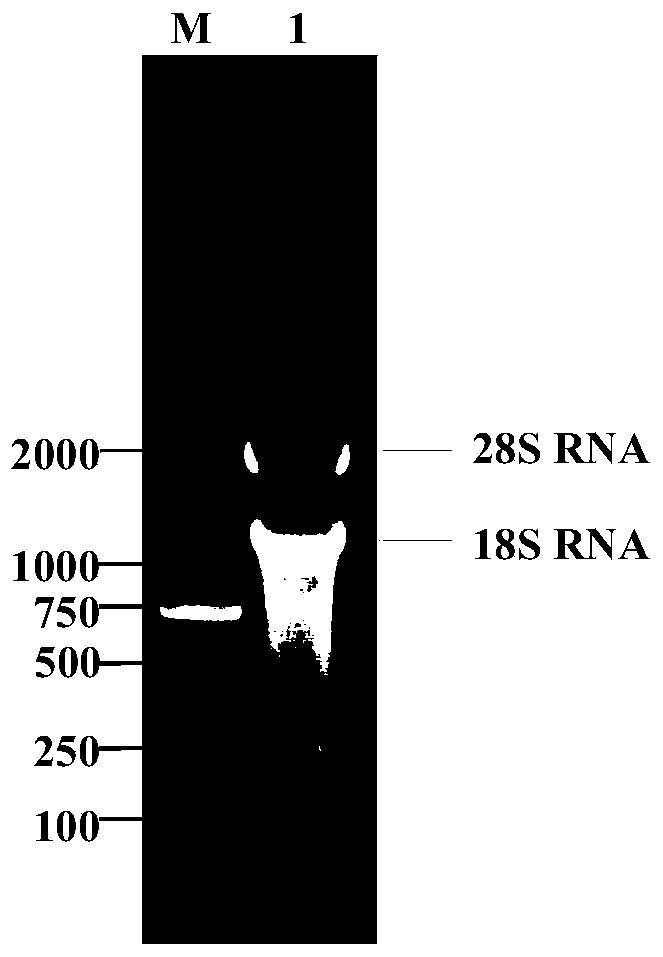
[0039] The Rhizopus oryzae (CICC40467) mycelium was ground with liquid nitrogen, and the total RNA was extracted with RNAiso plus Total RNA extraction reagent from Takara Company, and the genomic DNA was removed by DNase I treatment, agarose gel electrophoresis detection and absorbance analysis to ensure the extraction RNA does not degrade and contaminate genomic DNA.
[0040] The total RNA of Rhizopus oryzae extracted by the present invention is analyzed by absorbance: OD 260 / OD 280 =1.85, the RNA concentration is 89μg / ml, the agarose gel electrophoresis picture is as follows figure 1 According to the results of agarose gel electrophoresis, the 28S and 18S RNA bands in the extracted total RNA were clear and not degraded, indicating that the RNA integrity was good, and there was no genomic DNA contamination in the extracted RNA.
Embodiment 3
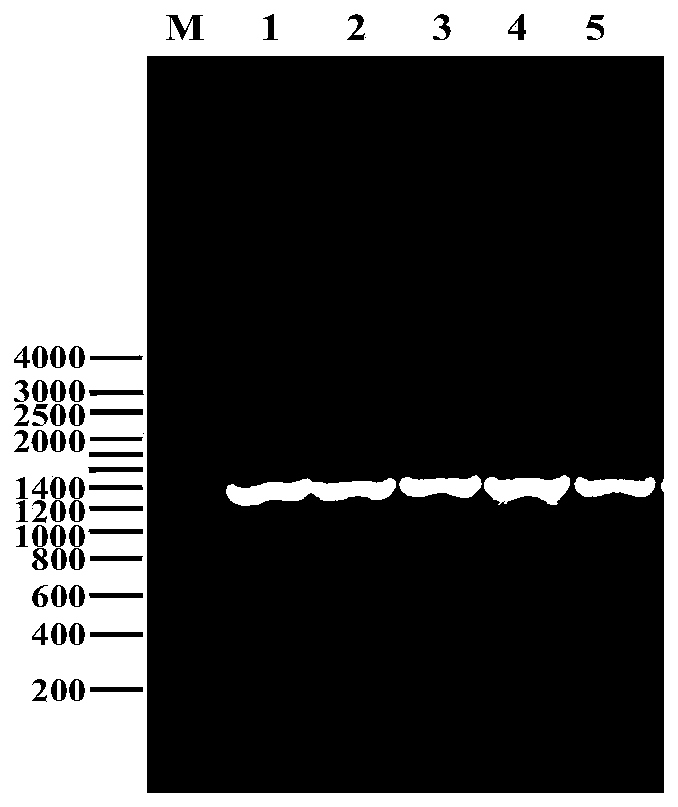
[0041] Embodiment 3: Rhizopus oryzae G6PDH gene clone
[0042] Using the extracted Rhizopus oryzae RNA as a template and primers F-g6pdh / R-g6pdh (see Table 1) for RT-PCR amplification to obtain the G6PDH gene fragment without introns, the PCR product agarose gel electrophoresis detection results like figure 2 , it can be seen from the figure that the band is between 1200 and 1400bp, which is consistent with the size of the coding sequence of the G6PDH gene (including intron 1439bp and exon 1346bp) reported in the database.
[0043] After purification and recovery, the amplified product was ligated with the PMD19-T Simple vector (purchased from TaKaDa) and then transformed. Picked 5 single colonies from the transformed plate for colony PCR. The results are as follows: image 3 . As can be seen from the figure, the selected 5 transformants were all positive. The No. 2 transformant was selected for expansion culture, the plasmid was extracted and verified by single and double...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com