Gene and protein related to lipid metabolism pathway of chlamydomonas reinhardtii, protein, gene mutation strains and applications thereof
A metabolic pathway, the technique of Chlamydomonas reinhardtii, applied in changing the oil content of Chlamydomonas reinhardtii, in the field of mutant algae strains, can solve the problems of slow progress, no new reports on synthesis and regulation pathways, and weaknesses
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Example 1. Cloning of lipid metabolism pathway-related genes (AT, TAGLP) in Chlamydomonas reinhardtii
[0082] The inventors found in the research that there are more than 2,500 genes in Chlamydomonas reinhardtii whose differential expression in the four growth stages is more than twice, and they play different roles in regulating the decomposition or synthesis of lipids. In the in-depth research, the inventor screened out genes related to the lipid metabolism pathway (such as AT, TAGLP, LPOGA, G3PDH, LPAAT and DAGAT six genes, the genes in the text are in italics, the same below), among which the AT-encoded protein AT is the acetyl transfer Enzyme, which can catalyze the reaction of acetyl coenzyme A and long-chain alcohol to form coenzyme A and long-chain ester; TAGLP encoding protein TAGLP is a triglyceride lipase, which can reduce triacylglycerol to diacylglycerol and fatty acid anion, from glycerol The reverse reaction for the synthesis of triacylglycerols. The pr...
Embodiment 2
[0085] Embodiment 2, the RNA (microRNA) that suppresses AT, TAGLP gene expression and the acquisition of coding sequence thereof, sequence analysis and the construction of AT, TAGLP gene silencing vector
[0086] 1. Acquisition of RNA (microRNA) for silencing AT gene and its reverse complementary sequence coding gene
[0087] According to the AT gene sequence, according to the principle of microRNA design (WMD3-Web MicroRNA Designer (http: / / wmd3.weigelworld.org / cgi-bin / webapp.cgi)), a target site was finally selected, which is located in the AT gene mRNA sequence transcription A sequence of 21bp in length downstream of the start site aug, the obtained microRNA that inhibits AT gene expression is named AT-miR, AT-miR: 5'-GAGUCGCAUAUGCUGUUAUUA-3', AT-miR acts on AT mRNA 96-115 position sequence 5'-GAGUCGCAUAUGCUGUUAUUA-3'. Then obtain its coding sequence and the coding gene capable of synthesizing the RNA (microRNA) and its reverse complementary sequence according to the AT-miR...
Embodiment 3
[0143] Embodiment 3, the acquisition of Chlamydomonas reinhardtii mutant strain AT-CR and TAGLP-CR
[0144] 1. Transforming AT and TAGLP gene silencing vectors into Chlamydomonas reinhardtii
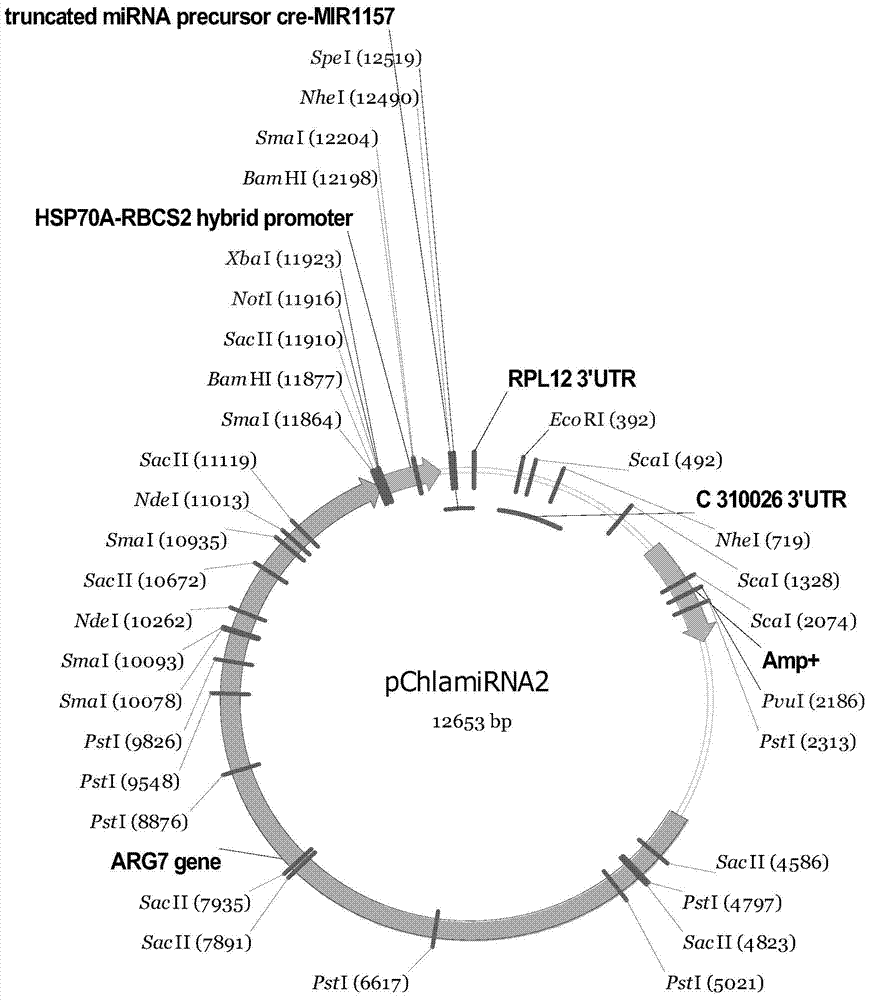
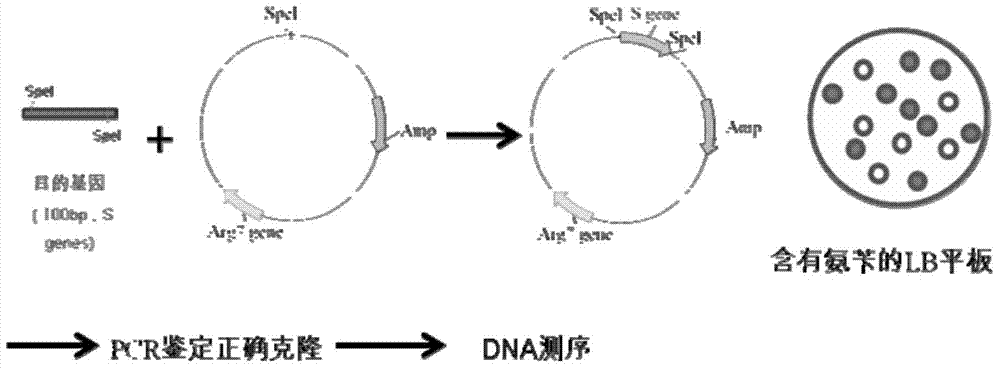
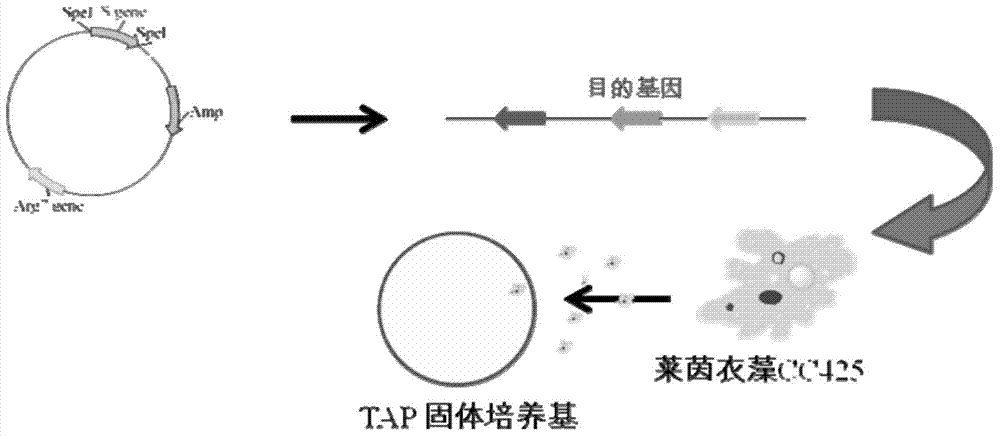
[0145] The AT gene silencing vector AT-mic-RNA2 carrying AT-miDNA and the TAGLP gene silencing vector TAGLP-mic-RNA2 carrying TAGLP-miDNA were transformed into Chlamdomonas reinhardtii CC425 and Chlamdomonas reinhardtii (from Beijing Life Sciences) respectively. Institute of Science) is an arginine auxotrophic algal strain that cannot synthesize arginine (Arg) by itself, while the silencing vectors AT-mic-RNA2 and TAGLP-mic-RNA2 corresponding to AT-miDNA and TAGLP-miDNA both contain Amp Resistance gene and Arg gene, so if a single clone grows on the arginine-free plate after transformation, it means that the transformation is successful. like image 3 As shown, the specific conversion method is as follows:
[0146] (1) Carry out single enzyme digestion of the plasmids AT-mic-RNA2 and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com