Multiple intermolecular rapid detection method for banana wilt bacteria and bacterial Erwinia carotovora
A technology of banana fusarium wilt and soft rot bacteria, applied in biochemical equipment and methods, microbial measurement/inspection, recombinant DNA technology, etc., can solve problems such as easy to miss farming time, labor and money consumption, time-consuming detection process, etc. , to achieve the effect of eliminating the spread of diseased seedlings and fast gene evolution rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
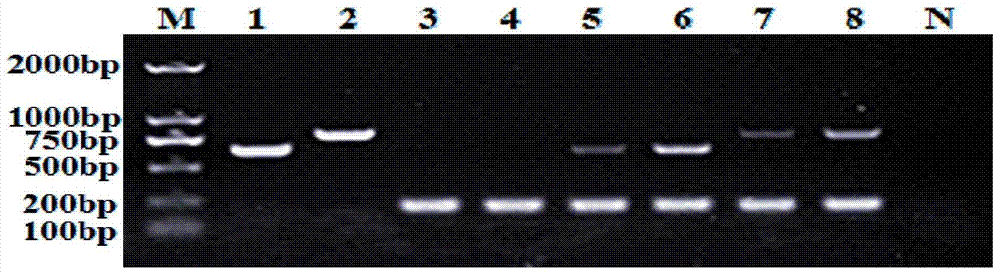
Image
Examples
Embodiment 1
[0065] Example 1: Design and Screening of Specific Primers for Physiological Race No. 1 and No. 4 of Banana Fusarium wilt
[0066] According to the complete genome sequences of Fusarium wilt No. 1 and No. 4 physiological races published by NCBI, search for gene sequences that may be related to the pathogenicity of Fusarium wilt of Banana Fusarium wilt and have insert / deletion differences between No. 1 and No. 4 physiological races . Sequence differential genes that may be related to pathogenicity were searched on Fusarium oxysporum f.sp.cubense race1contig438 (AMGP01000438.1) and Fusarium oxysporum f.sp.cubense race4contig195 (AMGQ01000195.1). See Seq1 and Seq2 for specific sequences. At 1515-1678bp of FOC4 (seq2), FOC4 has an insert fragment of about 160bp compared with FOC1; design suitable primers for extending the sequence on both sides of this fragment. The equivalent genome differential sequence can also be obtained by the following method: design appropriate primers ac...
Embodiment 2
[0068] Example 2: Design and screening of bacterial soft rot Dickeya sp. specific primers
[0069]Primers were designed based on the gyrB gene sequences of Dickeya bacteria that can cause bacterial soft rot and other genera, including Dickeya sp.MS1 (Genbank accession number: JQ284039), Dickeya dadantii3937 (Genbank accession number: NC_014500.1), Dickeya paradisiaca Ech703 (Genbank accession number: CP001654.1), Dickeya chrysanthemi NCPPB402 (Genbank accession number: CM001974.1), Dickeya dianthicola NCPPB3534 (Genbank accession number: NZ_CM001840.1), Pectobacterium carotovorum subsp.carotovorum PC1, CP001657.1), Pectobacterium wasabiae WPP163, CP001790.1), Pectobacterium atrosepticum SCRI1043, NC_004547.2), Escherichia coli (Escherichia coli P12b, CP002291. 1), Serratia marcescens WW4, CP003959.1, Citrobacter koseri ATCC BAA-895, CP000822.1, Serratia plymuthica AS9, CP002773.1, mouse Citrobacter rodentium ICC168, NC_013716.1, Shigella sonnei53G, NC_016822.1, Cronobacter sa...
Embodiment 3
[0071] Example 3: DNA Extraction and Specific Primer Identification of Banana Fusarium wilt No. 1 and No. 4 Physiological Race Strains
[0072] The strains of physiological race 1 and 4 of Fusarium wilt were preserved in 25% glycerol as spore suspension at -80°C. Inoculate the spore suspension onto the slant of the PDA test tube (potato dextrose agar medium), culture at 25°C for 7 days, then wash the surface of the cultured bacteria with 5mL sterile water to obtain the spore suspension, then draw 1mL of the spore suspension and add it to the liquid containing 150mL PDA Culture medium in a 250mL Erlenmeyer flask at 28°C and 150rpm for 3 days, and filter mycelium with 3 layers of lens-cleaning paper.
[0073] The extraction steps of fungal DNA are as follows:
[0074] a. take the mycelium and grind it into a fine powder with liquid nitrogen;
[0075] b. Take several 2mL centrifuge tubes, add 0.5mL volume of powder to each, then add 800μL FPCB solution (purchased from Sangon Bi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com