Method for producing 3-hydroxypropionic acid, acrylic acid and propanoic acid by immobilizing carbon dioxide in microorganisms
A technology of microorganisms and hydroxypropionic acid, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve problems such as high cost, consumption of petroleum resources, environmental pollution, etc., achieve fast growth and alleviate greenhouse benefits , the effect of reducing pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Example 1 Escherichia coli producing 3-hydroxypropionic acid
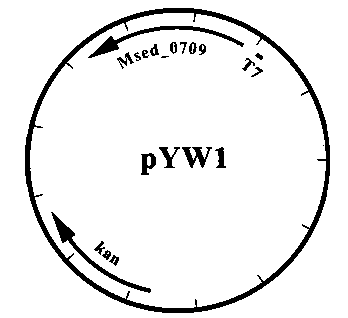
[0063] The Msed_1993 gene fragment on the plasmid pYW4 was used Xba I and Hin Digested with the dIII enzyme and connected to the back end of the Msed_0709 gene on pYW1 to construct a plasmid that simultaneously expresses the two genes Msed_0709 and Msed_1993, named pZL31, see appendix Figure 8 .
[0064] The plasmid pMSD8 constructed by Professor John Cronan's research group can overexpress acetyl-CoA carboxylase ACC in Escherichia coli (Davis M S, Solbiati J, Cronan J E. Overproduction of Acetyl-CoA Carboxylase Activity Increases the Rate of Fatty Acid Biosynthesis in Escherichia coli . Journal of Biological Chemistry, 2000, 275(37): 28593-28598.). In this example, this plasmid was used to increase the amount of malonyl-CoA.
[0065] Co-transform pMSD8 and pZL31 into Escherichia coli BL21 (DE3), use kanamycin and carbenicillin to screen the successful transformants, pick a single clone and culture i...
Embodiment 2
[0074] Example 2 Escherichia coli producing acrylic acid
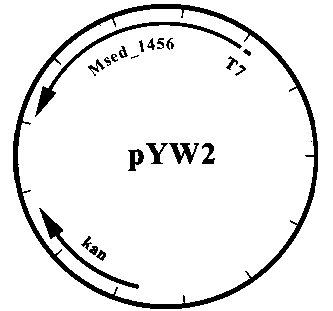
[0075] The Msed_1993 gene fragment on the plasmid pYW4 was used Xba I and Hin Digested with the dIII enzyme and connected to the back end of the Msed_2001 gene on pYW7 to construct a plasmid that simultaneously expresses four genes Msed_0709, Msed_1456, Msed_2001 and Msed_1993, named pZL32, see appendix Figure 9 .
[0076] The plasmid pMSD8 constructed by Professor John Cronan's research group can overexpress acetyl-CoA carboxylase ACC in Escherichia coli (Davis M S, Solbiati J, Cronan J E. Overproduction of Acetyl-CoA Carboxylase Activity Increases the Rate of Fatty Acid Biosynthesis in Escherichia coli . Journal of Biological Chemistry, 2000, 275(37): 28593-28598.) In this example, this plasmid was used to increase the amount of malonyl-CoA.
[0077] Co-transform pMSD8 and pZL32 into Escherichia coli BL21 (DE3), use kanamycin and carbenicillin to screen the successful transformants, pick a single clone and cul...
Embodiment 3
[0086] Example 3 Escherichia coli producing propionic acid
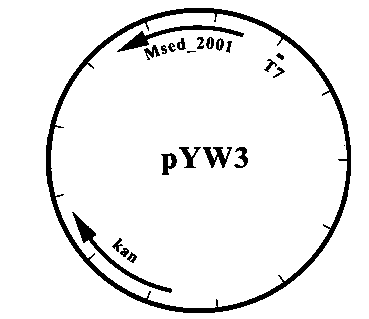
[0087] The Msed_1426 gene fragment on the plasmid pYW5 was used Xba I and xho After the I enzyme was cut off, it was connected to the back end of the Msed_2001 gene on pYW7, and a plasmid that simultaneously expressed the four genes Msed_0709, Msed_1456, Msed_2001 and Msed_1426 was constructed, named pZL33, see appendix Figure 10 .
[0088] The Msed_1993 gene fragment on the plasmid pYW4 was used Xba I and Hin Digested with the dIII enzyme and connected to the back end of the Msed_1426 gene on pZL33 to construct a plasmid that simultaneously expresses five genes Msed_0709, Msed_1456, Msed_2001, Msed_1426 and Msed_1993, named pZL34, see appendix Figure 11 .
[0089] The plasmid pMSD8 constructed by Professor John Cronan's research group can overexpress acetyl-CoA carboxylase ACC in Escherichia coli (Davis M S, Solbiati J, Cronan J E. Overproduction of Acetyl-CoA Carboxylase Activity Increases the Rate of Fa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com