Gene engineering bacterium capable of producing Kdo2-lipid A, construction method and applications thereof
A technology of genetically engineered bacteria and construction methods, applied in the field of Kdo2-lipidA-producing genetically engineered bacteria and its construction and application, can solve problems such as poor growth status, small final bacterial volume, unclear genetic background, etc., and achieve strain growth Good condition, single structure, beneficial to large-scale industrial production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1 Construction of mutant strain E.coliW3110△rfaD
[0051] 1. Obtaining the rfaD gene knockout fragment
[0052] The knockout fragment of the rfaD gene was obtained by chemical total synthesis or step-by-step PCR amplification, and its two ends were the upstream and downstream homology arms of the rfaD gene, and the middle was a kan fragment with an FRT site. The nucleotide sequence of the rfaD gene knockout fragment is shown in SEQ ID NO.1. The rfaD gene knockout fragment was cloned into pBlueScriptIISK(+) to obtain the recombinant plasmid pBlueScript IISK(+)-rfaDU-Fkan-rfaDD. Using the plasmid as a template, the knockout fragment rfaDU-Fkan-rfaDD can be amplified.
[0053] 2. Preparation and electrotransformation of knockout competent cells
[0054] Inoculate Escherichia coli with Red recombination helper plasmid pKD46 (DatsenkoKA, Wanner BL.One-Step inactivation of chromosome genes in Escherichia coli K-12using PCR products.Proc Natl Acad Sci USA,2000,97(12...
Embodiment 3
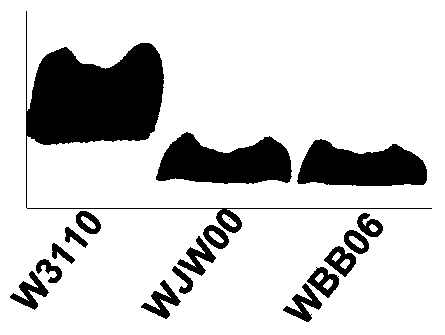
[0058] Example 3 Analysis of the lipopolysaccharide structure of the mutant strain E.coliΔrfaD
[0059] 1. Extraction of mutant strain E.coliΔrfaD lipopolysaccharide
[0060] LPS extraction of mutant strain E.coliΔrfaD, wild-type W3110 and WBB06 The extraction method of lipopolysaccharide adopts the classic thermal phenol method. Inoculate 5 mL of LB medium, culture overnight for 2 hours, and induce IPTG for 4 hours at OD=0.5). Take 1.5mL of bacterial liquid and centrifuge at 12000rpm for 3min to collect the bacterial cells. Thoroughly suspend the cells with 100 μL 1×TAEBuffer. Then add 200μL solution I (3gSDS; 0.6gTrizmabase (Tris); 6.4mL2MNaOH; 100mLddH2O), incubate at 55-60℃ for 70min or in boiling water bath for 15min. Then add an equal volume of phenol / chloroform at a volume ratio of 1:1 and mix well, centrifuge at 12000 rpm for 3 minutes to take the upper phase, and repeat 2 to 3 times. Add 200 μL ddH2O, 50 μL 3M NaAc (pH 5.2) and mix well, and precipitate LPS with 2...
Embodiment 4
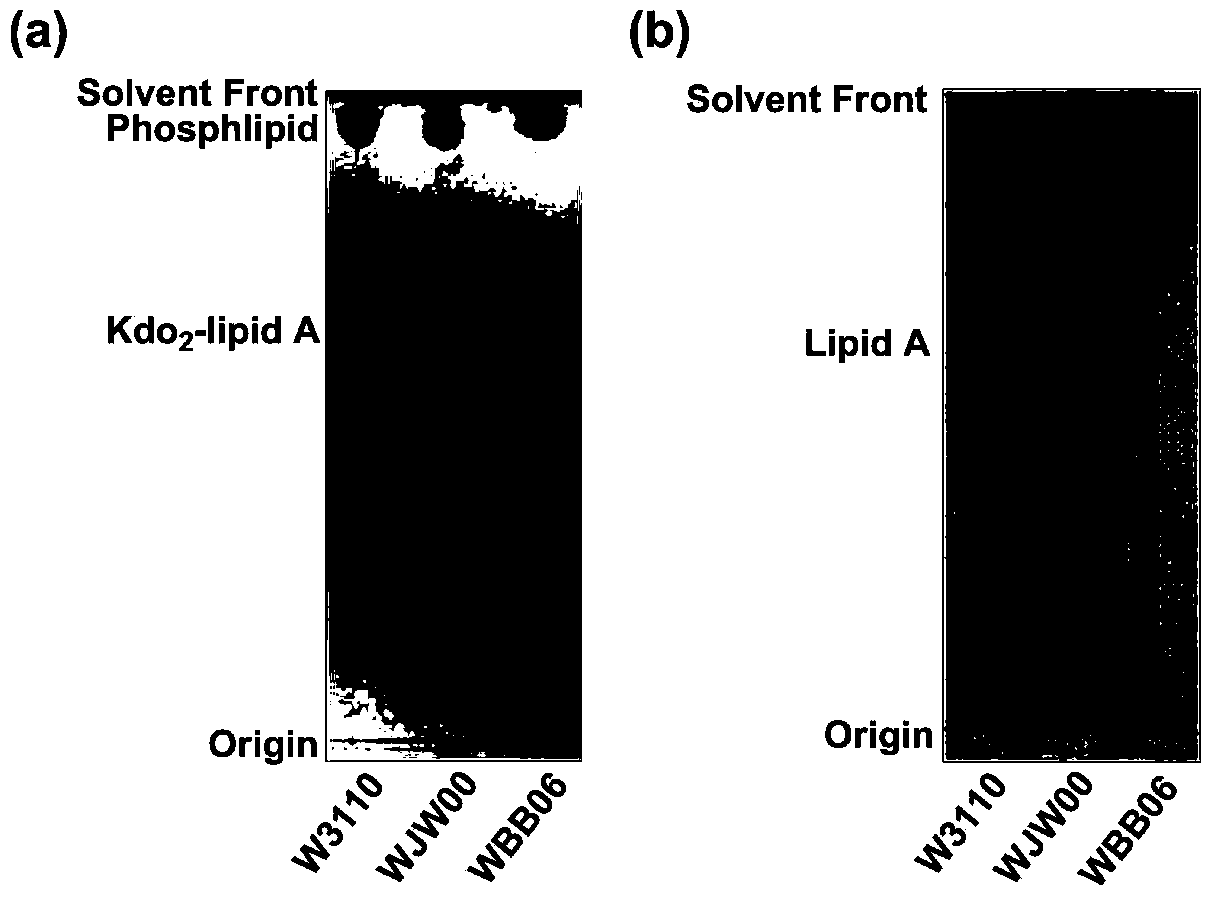
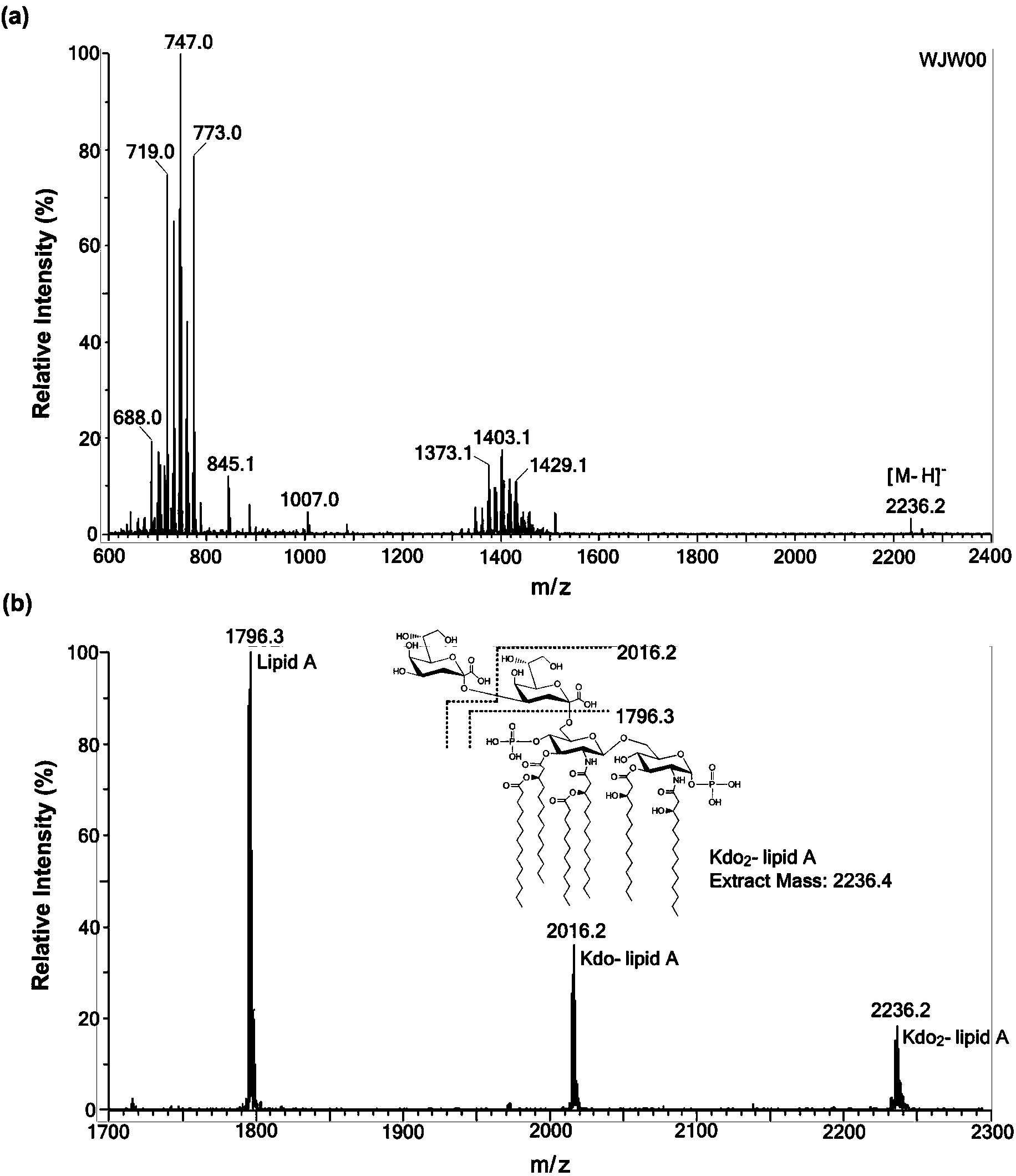
[0063] Example 4 Kdo of mutant strain E.coliΔrfaD 2 -Structure analysis of lipidA
[0064] 1. Kdo of mutant strain E.coliΔrfaD 2 -lipidA extraction and thin layer chromatography (TLC) analysis
[0065] Kdo 2-The extraction method of lipidA adopts the mixed phase extraction method of chloroform / methanol / 1.0M NaCl solution. The overnight cultured bacterial solution was divided into initial OD 600 =0.02 transferred to 200mLLB liquid medium, cultured to the late logarithmic phase of the bacteria at 37°C, centrifuged at 8000rpm for 10min to collect the bacteria, washed once with 0.1M NaCl solution, and then used Bligh-Dyer one-phase system (chloroform / methanol / 1.0M NaCl solution, 1:2:0.8, v / v / v) 76mL of suspended bacteria, magnetically stirred for 1h, centrifuged at 2000rpm for 30min to separate the phases, take the upper phase and add 20mL of chloroform and 20mL of 1.0MNaCl solution to make Bligh-Dyer two-phase System (chloroform / methanol / 1.0MNaCl solution, 2:2:1.8, v / v / v), ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com