A degenerate primer RT-PCR detection method for Eimeria virus
A technology of RT-PCR and degenerate primers, which is applied in the direction of biochemical equipment and methods, and the determination/inspection of microorganisms. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Primer design and synthesis
[0027] 1. Materials: Molecular biology software MEGA4, DNAMAN and NCBI network resources.
[0028] 2. Methods and results:
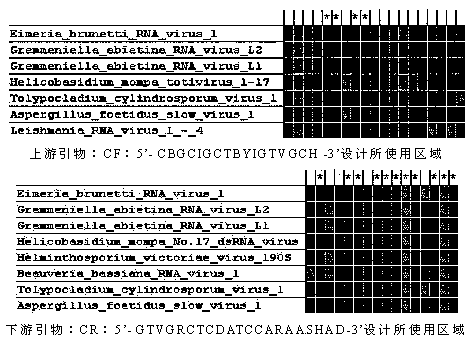
[0029] Sequence obtained: through Eimeria brucei virus ( E. brunetti RNA virus 1) Genome analysis, select the coat gene as the target sequence, and obtain the gene sequence from the GenBank public database, the accession number is: NC_002701. Use the coat protein sequence as the query sequence, and perform BlustP alignment on NCBI to obtain two highly conserved protein regions from 330 to 346 (IALGTAAAVAHCDPLVP) and 435 to 446 (PFFWIEPTTVLP), and compare the nucleic acid sequences of the corresponding positions of the rest of the reference sequence Separate downloads yielded Gremmeniella abietina RNA virus L2, Gremmeniella abietina RNA virus L1, Helicobasidium mompa totivirus 1-17, Helminthosporium victoriae virus, Beauveria bassiana RNA virus 1, Tolypocladium cylindrosporum virus 1, Aspergillus foetidus slowman vi...
Embodiment 2
[0035] Nucleic acid sample preparation
[0036] 1. Materials: Eimeria tenella, 2×STE buffer (Ph8.0), water-saturated phenol, chloroform, β-mercaptoethanol, 10% SDS, 70% ethanol, isopropanol, high-speed centrifuge .
[0037] 2. Methods and results:
[0038] 1) Broken Eimeria tenella oocysts: about 10 7 The coccidia to be tested were ground with liquid nitrogen, and dissolved in 1ml of 2×STE (pH8.0) buffer after grinding.
[0039] 2) Protein extraction: Add 30 μl of β-mercaptoethanol, 0.6 ml of water-saturated phenol, 0.4 ml of chloroform, and 140 μl of 10% SDS to the above coccidia solution. Thoroughly homogenize for 5 minutes. Centrifuge at 12000r / min for 5 minutes at 4°C, and take the supernatant.
[0040] 3) Acquisition of total nucleic acid: mix the supernatant with an equal volume of cold isopropanol, let stand at -80°C for 10 minutes, centrifuge at 15,000r / min for 15 minutes at 4°C, wash the pellet with 70% cold ethanol solution, and store at 4°C Under conditions, c...
Embodiment 3
[0044] Detection of Eimeria tenella virus by RT-PCR with degenerate primers
[0045] 1. Materials: Eimeria tenella RNA crude extract, PrimeScript™ Reverse Transcriptase (2680Q) reverse transcriptase kit, TaKaRa LA Taq (RRO2MQ) nucleic acid polymerase kit, agarose, ethidium bromide solution, Biorad PCR instrument.
[0046] 2. Methods and results:
[0047] 1) 1st-Stand cDNA synthesis: Prepare the following mixture in Microtube:
[0048]
[0049] Denature at 99°C for 1 minute, and immediately transfer to -40°C for quick freezing.
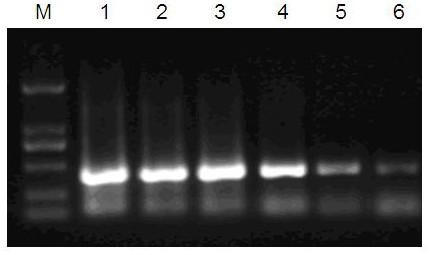
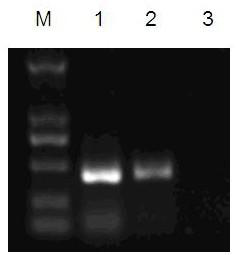
[0050] The amount of template RNA was used for gradient experiments, which were divided into six gradients, 50ng / μl, 5ng / μl, 0.5ng / μl, 0.05ng / μl, 0.005ng / μl, 0.0005ng / μl. When the template RNA concentration is 0.05ng / μl, PCR results can still be clearly interpreted. (see attached figure 2 )
[0051] Add the following components in the above reaction solution:
[0052]
[0053] Reaction at 30°C for 10 minutes, reaction at 42°C for 45 minut...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com