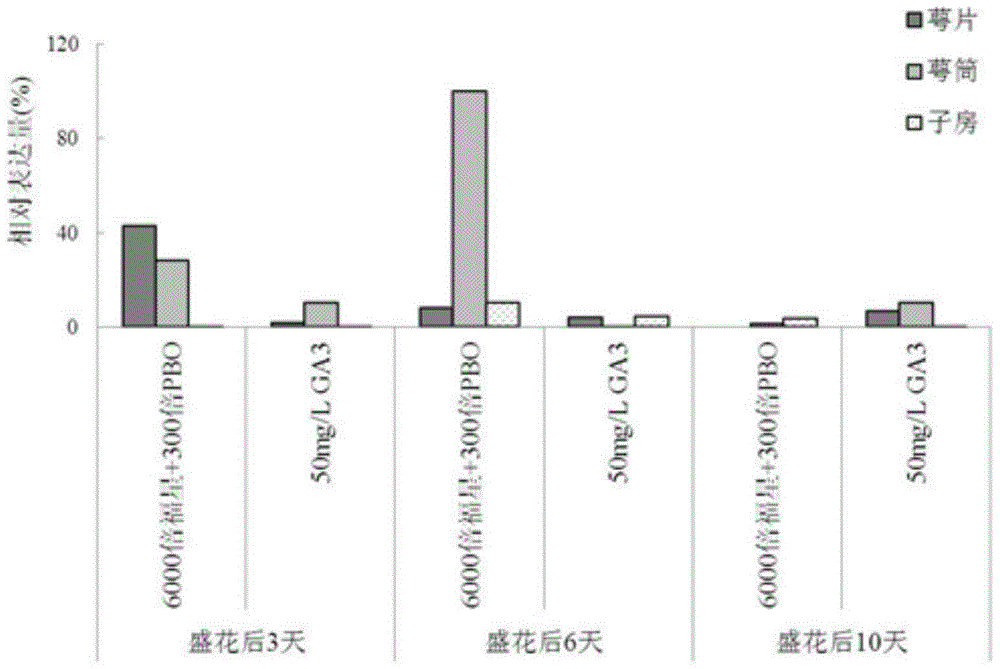
Pear transcription factor psjointless and its application
A transcription factor and gene technology, applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve the problems of functions that need to be further verified and limited research, and achieve the effect of achieving environmental friendliness and reducing agricultural costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 PsJOINTLESS gene isolation, cloning and expression analysis
[0035] RNA was extracted from the calyx of'Korla Fragrant Pear', and the first strand cDNA obtained by reverse transcription was used to amplify the full length of the PsJOINTLESS gene.
[0036] The RNA was extracted using the Trizol kit (purchased from TakaRa, operated in accordance with the operating instructions provided by the kit), and 1 μg of total RNA sample was extracted with 1 U DNase (purchased from Thermo). The first strand cDNA synthesis uses TOYOBO reverse transcription kit (operate according to the instructions provided by the kit). The primer pairs for the amplified gene are: PsJOINTLESS-F1: 5'-ATGGATGGCGAGGGAGAAAATTCAG-3' (SEQ ID No. 3); PsJOINTLESS-R1: 5'-GGTCACCTTATCAGATGACTTAAACGCAC-3' (SEQ ID No. 4). The 50 μL reaction system includes 200 ng cDNA, 1× buffer (TransStart FastPfu Buffer), 10 mM dNTP, 1 U Taq polymerase (TransStart FastPfu DNA Polymerase) (the aforementioned buffer and T...
Embodiment 2
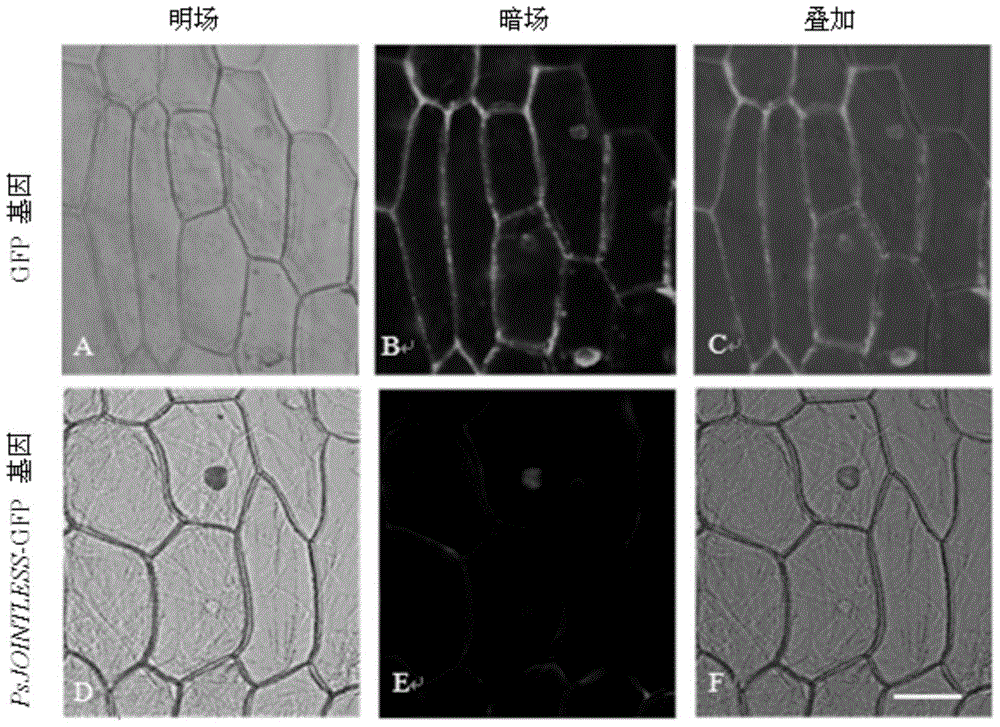
[0039] Example 2 Subcellular localization of PsJOINTLESS gene
[0040] Since PsJOINTLESS gene has a nuclear localization signal (NLS), this study used Agrobacterium to infect onion epidermis to study the subcellular localization of PsJOINTLESS gene. The entire ORF (reading frame) of the PsJOINTLESS gene was amplified by RT-PCR, and Nco I and Spe I were added at both ends of the amplification primers. Firstly, the amplified product is loaded on the pMD19-T vector to obtain a pMD19-TN / S-PsJOINTLESS recombinant vector. At the same time, pMD19-TN / S-PsJOINTLESS and pCAMBIA1302 were digested with Nco I and Spe I, and the recovered genes were connected with the vector backbone to obtain the pCAMBIA-1302-PsJOINTLESS-GFP recombinant plasmid. The recombinant plasmid was double-enzymed Digestion identification, the correct recombinant plasmid identified by double enzyme digestion was sequenced, and the results showed that the obtained recombinant plasmid pCAMBIA-1302-PsJOINTLESS-GFP was co...
Embodiment 3
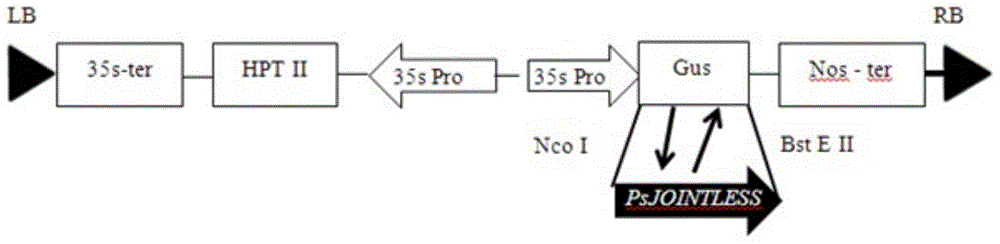
[0041] Example 3 Construction of plant transformation overexpression vector
[0042] According to the analysis of the multiple cloning site of pCAMBIA-1301 vector and the restriction site analysis on the coding region sequence of PsJOINTLESS gene, Nco I and BstE II were selected as endonucleases. According to the general principle of designing primers, use PrimerPrimer3.0 software to design primers with restriction sites. The primer pair sequence is as follows:
[0043] PsJOINTLESS-F3:TG CCATGG TGATGGCGAGGGAGAAAATTCAG (SEQ ID NO. 7)
[0044] PsJOINTLESS-R3:GG GGTCACC TTATCAGATGACTTAAACGCAC (SEQ ID NO. 8)
[0045] Using the correctly sequenced preserved bacterial solution to extract the plasmid as a template, clone the gene containing restriction enzyme sites. The annealing temperature for PCR amplification is 52°C, and the PCR reaction system and amplification procedure are the same as in Example 1. Recover the band of interest and connect it to the pMD19-T vector to construct a ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com