Pseudorabies virus gene-deleted attenuated strain as well as preparation method and application thereof
A pseudorabies virus and attenuated strain technology, applied in the field of bioengineering, can solve the problem of poor protection effect of mutant strains
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
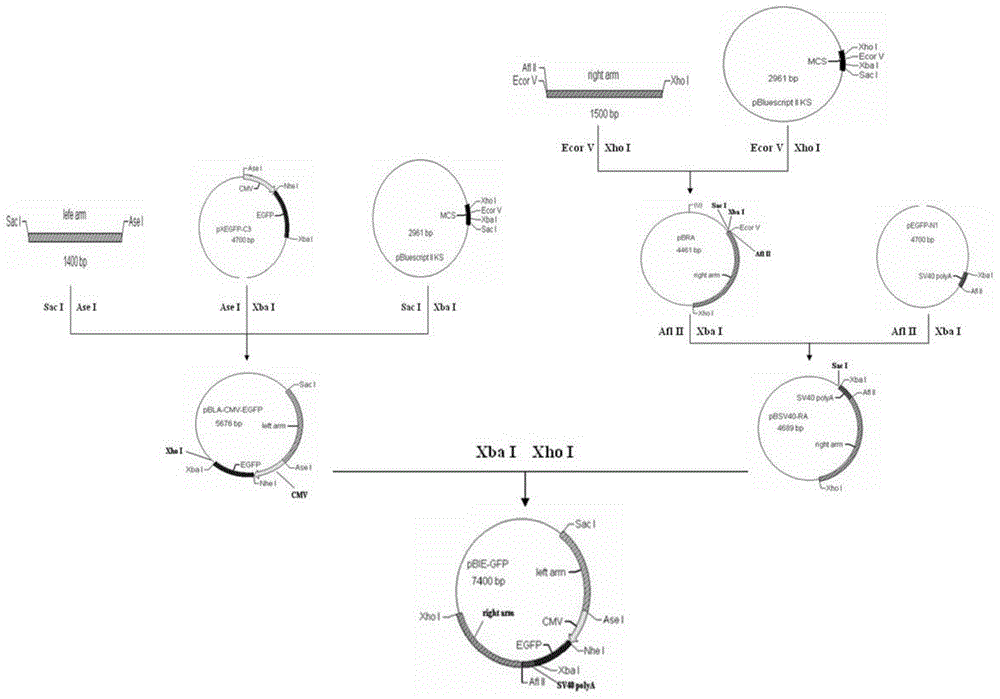
[0051] Construction of embodiment 1 PRV JS-2012gI / gE gene deletion transfer vector pBIE-GFP
[0052] Through the PCR method, two fragments located at bases 121632 to 123031 and bases 124854 to 126372 in the PRV JS-2012 genome were amplified and used as the left and right recombination arms of homologous recombination. At the same time, cut out the CMV promoter-EGFP gene fragment from the pEGFP-C3 plasmid by endonuclease digestion, cut out the SV40polyA signal sequence fragment from the pEGFP-N1 plasmid, and combine the left recombinant arm-CMV promoter-EGFP gene -SV40polyA signal sequence-right recombination arm was sequentially assembled into the pBluescript SK(+) vector to obtain the recombination transfer vector pBIE-GFP. The specific construction process is as follows (see figure 1 ):
[0053] 1.1 Primer design
[0054] According to the whole genome sequence of PRV JS-2012 strain, 2 pairs of primers PRELF / PRELR and PRERF / PRERR (see Table 1) were designed with Oligo6.0 s...
Embodiment 2
[0078] Construction of embodiment 2 PRVJS-2012-△gI / gE-EGFP strain
[0079] The cells were infected with PRVJS-2012 and the total DNA was extracted, and the total DNA was co-transfected with pBIE-GFP to obtain the recombinant virus. After 6 rounds of plaque purification, the recombinant virus JS-2012-△gI / gE-EGFP with deletion of gI and gE genes and EGFP marker was obtained.
[0080] 2.1 Primer design
[0081] According to the whole genome sequence of PRVJS-2012 strain, two pairs of primers gIU / gID and gEU / gED were designed with Oligo6.0 software to amplify the full-length sequence of gI and gE of PRV JS-2012. PRV commonly used detection primers gBup / gBdown, and identification primers gEup / gEdown for distinguishing wild virus from vaccine virus are kept by our laboratory. The primer sequences are listed in Table 2.
[0082] Table 2 PRV detection primers
[0083]
[0084] 2.2 Extraction of PRV JS-2012 genome
[0085] Inoculate the PRV JS-2012 strain into confluent monolay...
Embodiment 3
[0092] Example 3 Deletion of PRVJS-2012-△gI / gE-EGFP virus marker gene
[0093] Referring to the ideas in Example 1, the Ase I restriction site at the end of the left recombination arm was replaced with Xba I. Put the left recombination arm into the pBRA plasmid constructed in 1.5 to form a new homologous recombination transfer vector pBIE (see Figure 13 ). The cells were infected with PRV JS-2012-△gI / gE-EGFP and the total DNA was extracted, and the total DNA was co-transfected with pBIE to obtain the recombinant virus. After three rounds of plaque purification, the recombinant virus PRV JS-2012-△gI / gE without EGFP marker was obtained.
[0094] 3.1 Primers
[0095] Since the experiment needs to replace the Ase I restriction site at the end of the left recombination arm with Xba I in Example 1, a downstream primer needs to be redesigned to replace the Ase I restriction site with Xba I. The sequence of the primer PRELR-X is: 5'TTCTAGAAACTAGGGTCCACGACGCGCAGGCTG3' (SEQ ID NO.2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com