Helianthus tuberosus fructan 1-exo-hydrolase gene Ht1-FEH II and application thereof
A technology of fructan and hydrolase, which is applied in the field of genetic engineering and can solve the problems of no public reports of amino acid or nucleotide sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
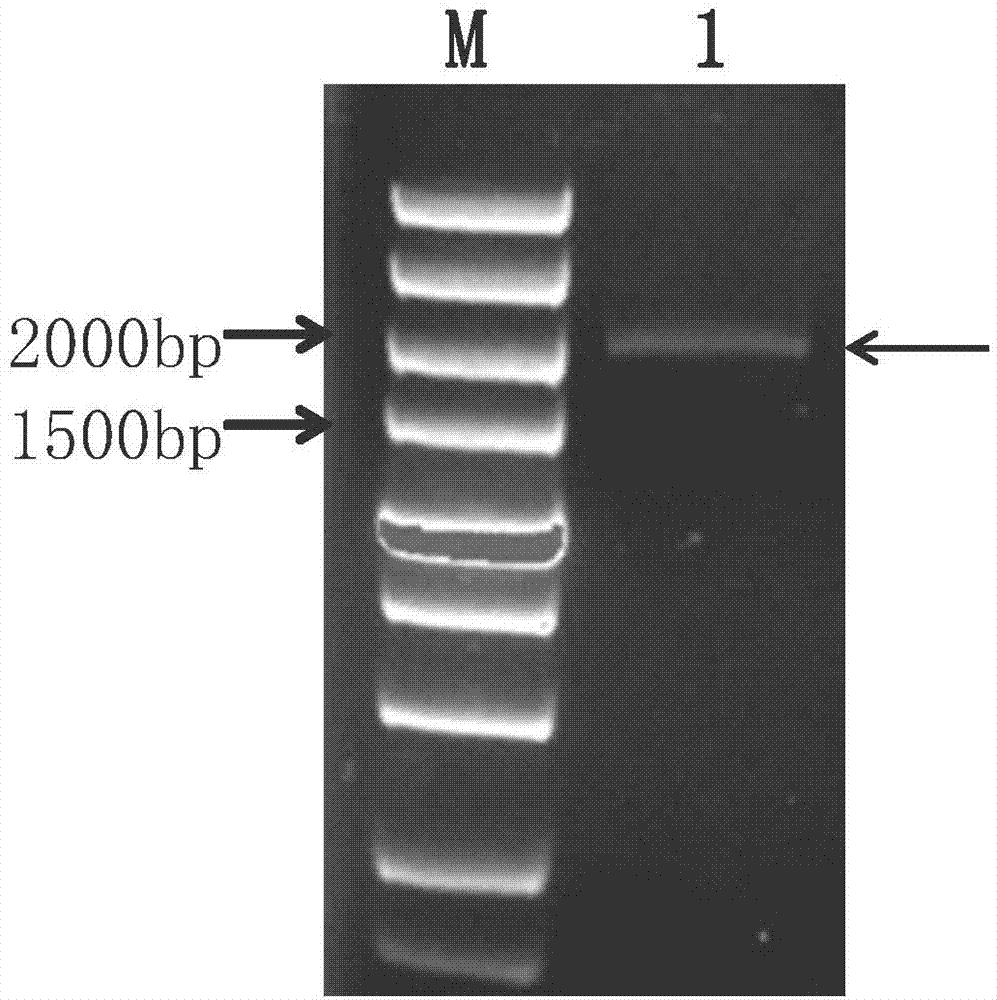
[0031] The cloning of embodiment 1Ht1-FEH II gene
[0032] The young leaves of the Jerusalem artichoke variety "Nanyu No. 1" at the 4th leaf stage were taken, and the total plant RNA was extracted according to the instructions of the OMEGA plant RNA extraction kit, and the cDNA template was obtained by two-step reverse transcription with the TaKaRa reverse transcription kit.
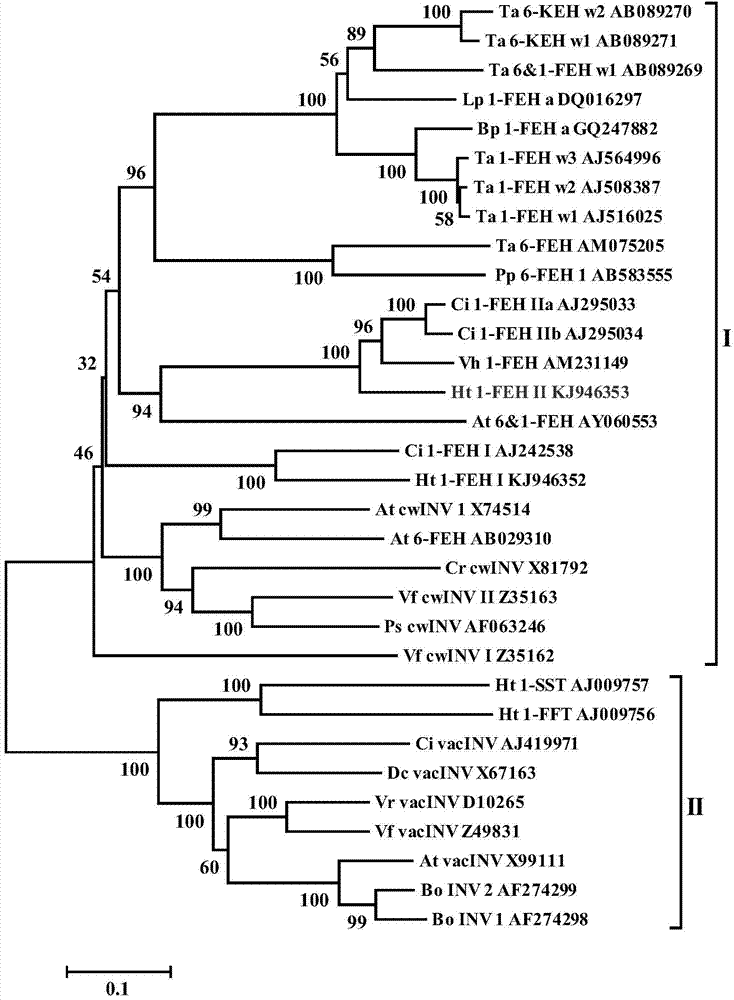
[0033] Blast the existing 1-FEH gene coding sequence (coding sequence, CDS) sequence of other species into Jerusalem artichoke (NCBI) and sunflower EST database (http: / / compbio.dfci.harvard.edu / tgi / plant.html), The EST sequence with high similarity was screened and spliced with DNAstar software to obtain an EST sequence with a full length of 2061bp. The EST sequence was compared with the existing 1-FEH gene CDS sequence and found that the EST sequence was similar to the Ci1-FEH of chicory. The CDS sequence similarity of IIa (AJ295033) is 80.3%, and that of Ci1-FEH IIb (AJ295034) is 79.6%, and there are...
Embodiment 2
[0048] Example 2 Construction of pPICZαC-picFEH II expression vector
[0049] According to the sequence analysis results of the fructan exohydrolase gene Ht1-FEH II, the upstream primer removed the predicted N-terminal signal peptide sequence: MNQLLSSFLTLCFLVLVFDTPTSNA, a total of 25 amino acids, starting from the 26th amino acid to design the upstream primer, and adding Xho I Restriction site: C|TCGAG, plus Kex2 signal cleavage sequence: AAAAGA, plus protective base: CCG, using the yeast α factor leader peptide sequence that comes with the vector as the signal peptide, construct a secreted expression vector, so that Recombinant fructan exohydrolase can be secreted into the extracellular of Pichia pastoris. The downstream primer removes the stop codon of the sequence itself, and adds a fusion 6×His tag after the expression frame to make the recombinant protein a fusion protein, which is convenient for purification and detection after protein expression, plus an Xba I restricti...
Embodiment 3
[0057] Embodiment 3 Expression vector transforms Pichia pastoris
[0058] (1) Linearization of plasmids
[0059] The restriction endonuclease SacI was used to linearize the vector pPICZαC-picFEH II, so that the linearized plasmid DNA could be integrated with the host genome DNA through homologous recombination.
[0060] The digestion system is as follows: 10×NEB Buffer 415 μL, DNA≤6 μg 50 μL, 100×BSA 1.5 μL, Sac I 3 μL, ddH 2 O 80.5 μL, total volume 150 μL.
[0061] Enzyme digestion at 37°C overnight, 1% agarose gel electrophoresis to detect the linearization effect, and use a nucleic acid recovery kit (PCR clean Kit) to recover the linearized product.
[0062] (2) Competent cells of Pichia pastoris X-33 (INVITROGEN company) were prepared.
[0063] (3) Electrotransform Pichia pastoris X-33 with the linearized product prepared in step (1), and obtain positive transformants through YPDS+ZeocinTM (100 μg / mL) resistance plate screening.
[0064] ⑷ Detection of positive clones ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com