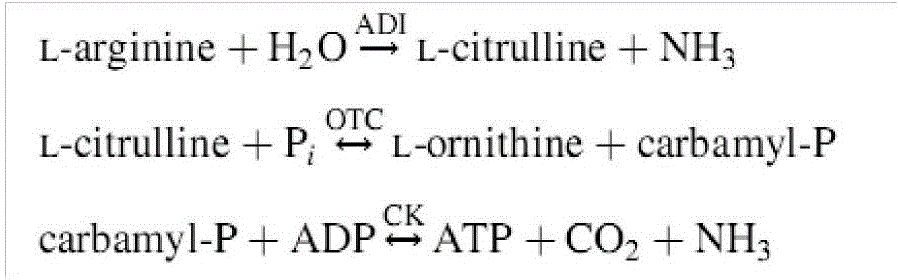Method for efficiently screening lactic acid bacteria capable of sufficiently utilizing citrulline
A high-efficiency technology of citrulline, applied in the field of microorganisms, can solve the problems that there are no application examples of citrulline, and achieve the effect of reducing the content
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Cultivable microorganism isolation in embodiment 1 soy sauce
[0025] Sampling and dilution from the finished koji of brewed soy sauce were applied to MRS and nutrient broth medium plates respectively, and enriched and cultivated at 37°C for 5 days. Select a single colony with different colony forms and inoculate it on a medium plate containing MRS and nutrient broth, and separate it by streaking for pure culture. Four days later, the pure culture was inoculated into MRS and nutrient broth liquid medium and cultured for four days to enrich the cells and extract the genome. The 16S rDNA was obtained by PCR with common primers for 16S rDNA of bacteria, and the 16S rDNA sequencing results were compared with the database.
[0026] The deposit number of Pediococcus acidilacticiBBE 1120 is CCTCC NO:M 2013732.
[0027] Tetragenococcus halophilus BBE R23 was deposited in China Center for Type Culture Collection (CCTCC), Wuhan, China, Wuhan University on October 20, 2013, and ...
Embodiment 2AD
[0029] Embodiment 2ADI pathway gene analysis
[0030]According to the data published in NCBI GenBank about Weissella, Staphylococcus, and lactobacillus: Weissella confusa LBAE C39-2 GenBank Assembly ID: GCA_000239955.2, Weissella cibaria KACC 11862 )GenBank Assembly ID: GCA_000193635.2, Staphylococcus aureus subsp.aureusNCTC 8325) GenBank Assembly ID: GCA_000013425.1, Pediococcus acidilactici DSM 20284) GenBank Assembly ID: halophilicum 1 (GCA_03 Tetragenococcus halophilus NBRC 12172) GenBank: AP012046.1, the analysis shows that the key genes of the ADI pathway of Weissella, Staphylococcus, and Lactobacillus only have a single copy, that is, only three genes, arcA, arcB, and arcC. The T.halophilus NBRC12172 pathway consists of arcA, arcB, arcC genes on arc operon and two additional copies of arcB, arcC genes other than arc operon, that is, arcA, arcB, arcC, arcB1, arcC1, arcB2 , arcC2 these 7 genes. The copy number of this ADI pathway gene may be related to the strain's abil...
Embodiment 3
[0031] Example 3 Analysis and Validation of ADI Pathway Gene Clusters of Tetradynococcus halophilus
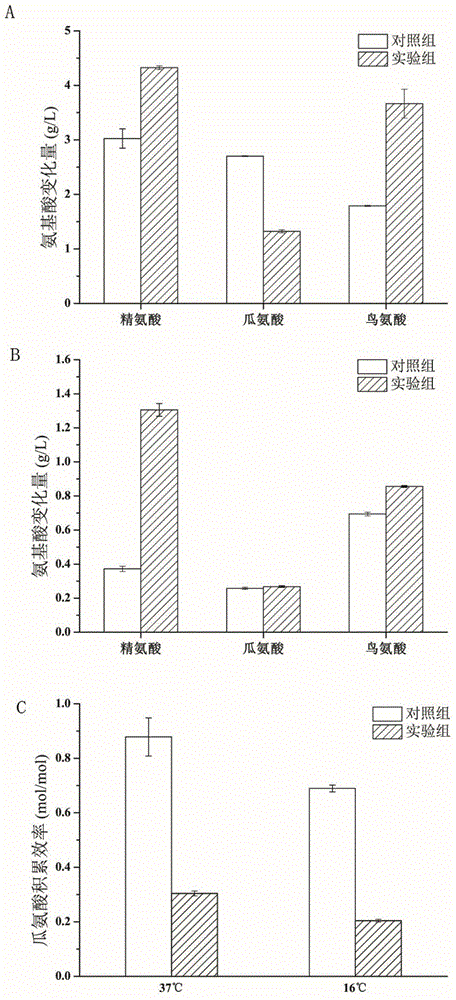
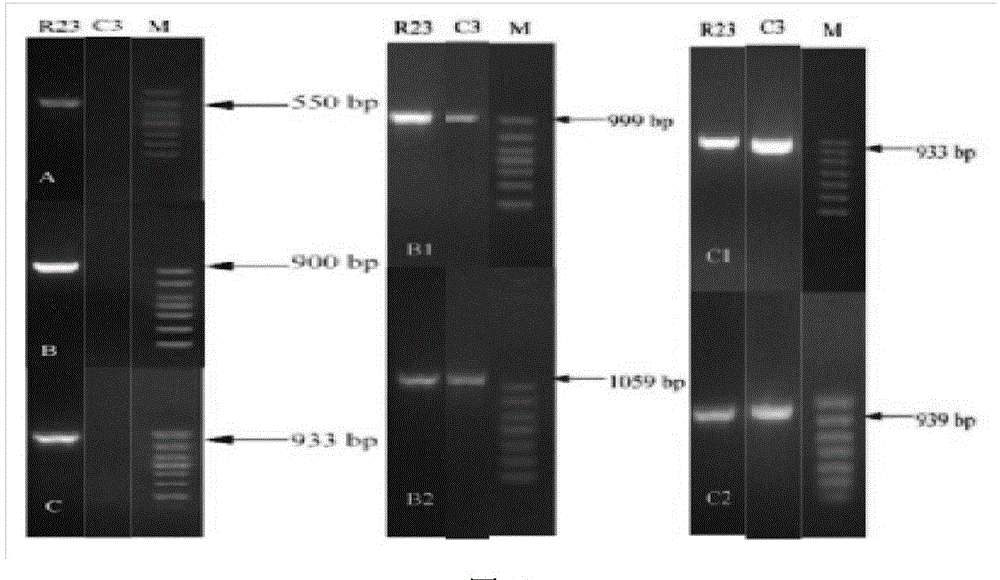
[0032] Pick Tetragenococcus halophilus BBE R23 (CCTCC NO:M 2013480) and Tetragenococcus halophilus BBE C3 (CCTCC NO:M 2013479) obtained from koji and preserved in glycerol tubes at -80°C, and mark on 10% NaCl MRS solid medium Wire. After culturing at 30°C for four days, pick a single colony and perform colony PCR. The primers are shown in Table 1 to verify whether there are arcA, arcB, arcC, arcB1, arcC1, arcB2, arcC2 genes. The PCR results showed that there was a complete arc operon (arcA, arcB, arcC) and two extra copies of arcB gene and arcC gene in the genome of T. halophilus R23; there was not a complete arc operon in the genome of T. halophilus C3 ( Deletion of arcA, arcB, arcC), but there are two copies of arcB1, arcC1 genes other than arc operon.
[0033] The primers used in the embodiment of table 1
[0034]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com