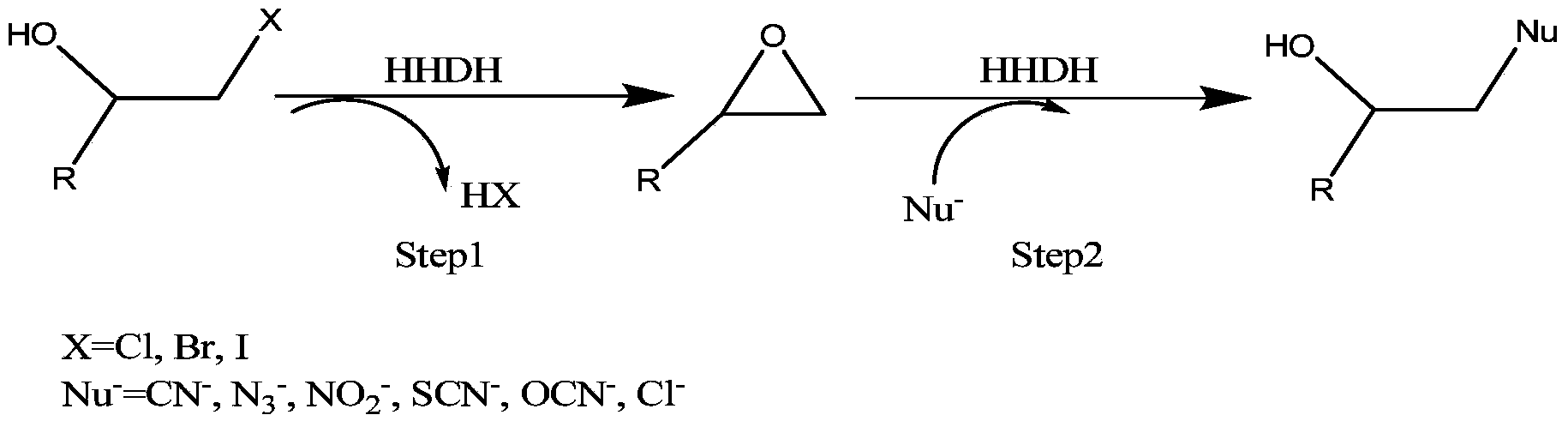
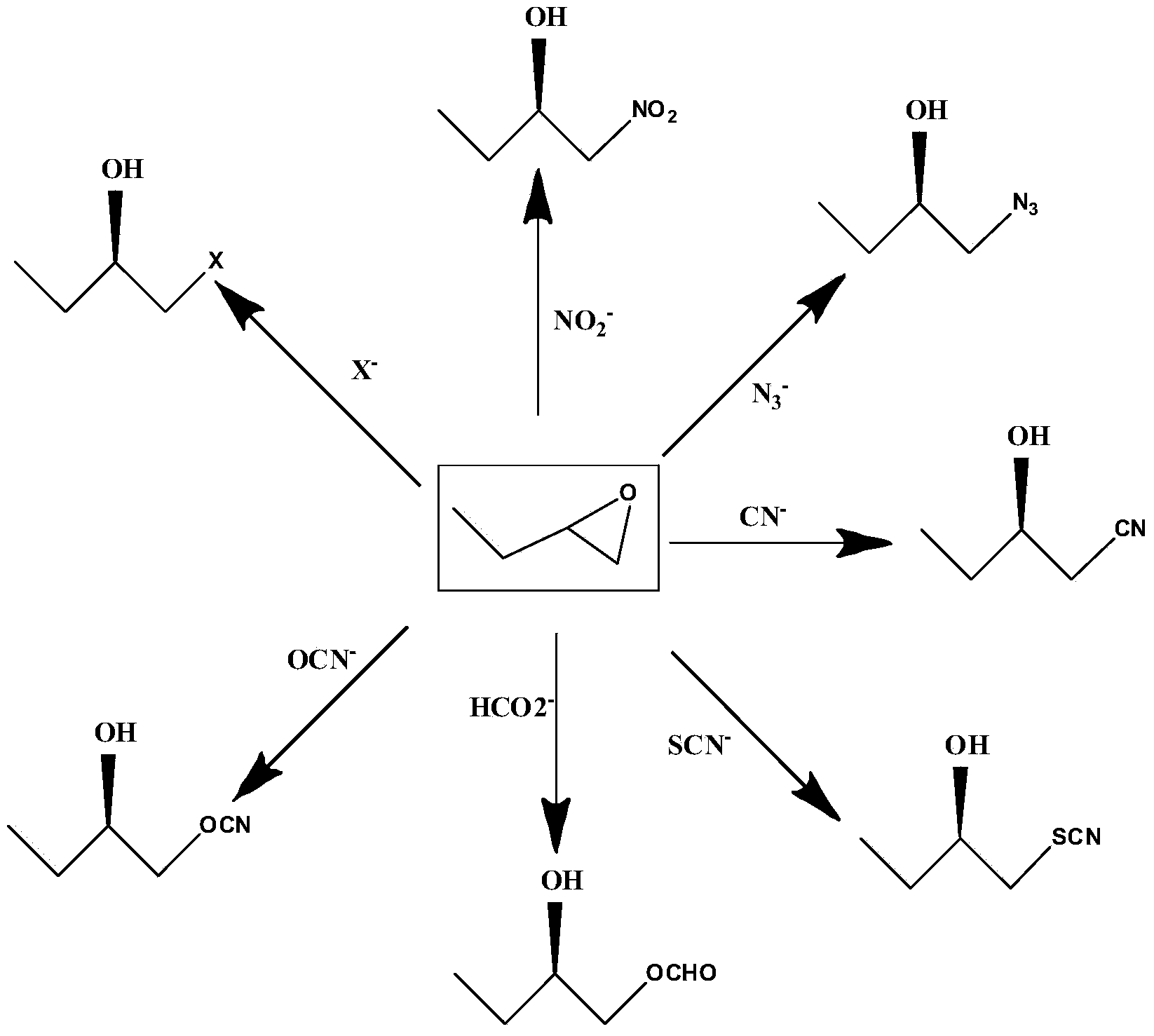
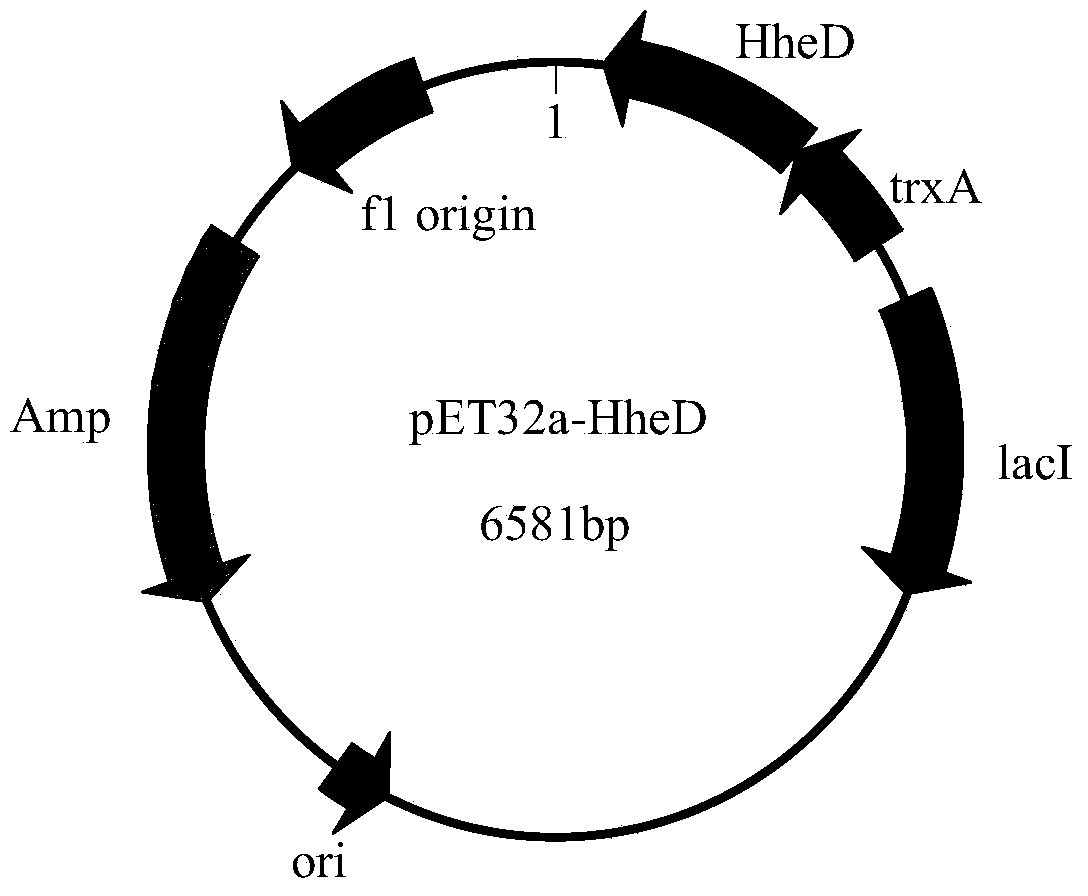
Parvibaculum lavamentivorans ZJB 14001, halohydrin dehalogenase enzyme gene, enzyme, engineered bacterium and application
A technology of halohydrin dehalogenase and genetically engineered bacteria, which can be used in genetic engineering, applications, bacteria, etc., and can solve problems such as limiting the application of halohydrin dehalogenase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Screening and Identification of Example 1 Bacterial Strains
[0049] The soil and seawater from the coastal area of Zhoushan were collected, enriched with the enrichment medium, spread on the plate of the enrichment medium, and cultured at 30°C for one week. The single bacterium colony of the different form that separates obtains with fermentation medium (composition of fermentation medium: NaCl 5g / L, beef extract 10g / L, peptone 5g / L, yeast powder 5g / L, solvent is deionized water, pH value natural .) Cultivated at 30°C to obtain a large amount of wet bacteria.
[0050] Using 0.2g of wet bacteria as a catalyst, 20mmol / L 1,3-dichloropropanol as a substrate, and 20ml of 50mM, pH7.0 PBS as a reaction medium, the conversion reaction was carried out at 37°C for 30min. After the reaction After the reaction solution was extracted with ethyl acetate, the ethyl acetate layer was taken to analyze the content of the product in the reaction solution by GC. A strain with halohydr...
Embodiment 2
[0055] The culture condition of embodiment 2 food cleaning agent parvobacterium ZJB14001
[0056] (1) Incline cultivation
[0057] Inoculate the slant culture medium with Escherichia coli ZJB14001 and culture it at 30°C for 7 days to obtain slant bacteria;
[0058] The composition of the slant medium is: NaCl 5g / L, peptone 10g / L, CaCl 2 0.02g / L and agar 2.0g / L, the solvent is deionized water, and the pH value is natural.
[0059] (2) Fermentation culture
[0060] Bacterial colonies were picked from the slant and inoculated into the fermentation medium, and cultured at 30° C. and 150 rpm for 5 days to obtain a bacterium-containing fermentation broth.
[0061] The components of the fermentation medium are: NaCl 5g / L, peptone 10g / L, CaCl 2 0.02g / L, the solvent is water, and the pH value is natural.
Embodiment 3
[0062] Embodiment 3: the cloning of halohydrin dehalogenase HHDH-PL gene
[0063] The total genomic DNA of the Parvibaculum lavamentivorans ZJB14001 cells obtained in Example 2 was extracted with a rapid nucleic acid extractor (purchased from eppendorf), and PCR was performed using the genomic DNA as a template and primers 3 and 4 as amplification primers. The amount of each component in the PCR reaction system (total volume 100 μL): 10×Pfu DNA Polymerase Buffer 10 μL (Mg 2+ ), 0.5 μL of 10 mM dNTP mixture (2.5 mM each of dATP, dCTP, dGTP, and dTTP), 0.5 μL of each cloning primer 3 and primer 4 at a concentration of 50 μM, 1 μL of genomic DNA, 1 μL of Pfu Taq DNA Polymerase, and 86.5 μL of nucleic acid-free water.
[0064] Primer 3: 3'- CCATGG CGCGCAGCATTCTC-5′;
[0065] Primer 4: 3'- CTCGAG CTAGCGCGCGGTGGCCC-5′
[0066] The PCR instrument of Biorad was used, and the PCR reaction conditions were: pre-denaturation at 94°C for 3min, followed by a temperature cycle of 94°C ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com