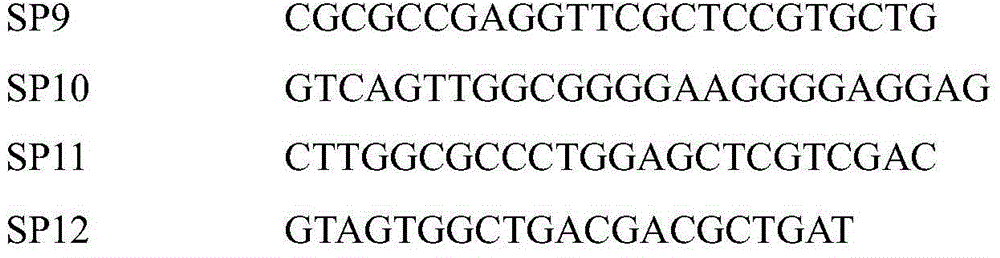Tyrosinase coding gene melC derived from streptomyces kathirae SC-1 and proteins of tyrosinase coding genes melC
A tyrosinase gene, tyrosinase technology, applied in genetic engineering, plant gene improvement, DNA/RNA fragments, etc., can solve the problems of expensive and large-scale production and application of melanin, cumbersome process and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Embodiment 1: (1) Streptomyces kathirae SC-1 is inoculated in culture medium (g / L: glucose 1, peptone 10, NaCl 5, CaCl 2 0.1, pH 6.2) 36h, 8000g centrifugation to collect the fermentation broth to remove mycelia.
[0030] (2) Under the condition of ice bath, add ammonium sulfate to the fermentation broth until its saturation is 65% to precipitate tyrosinase protein, continue to stir the fermentation broth for 20 minutes, centrifuge at 12000g for 1 hour, discard the supernatant carefully, and collect the protein For precipitation, use Buffer A to resuspend the enzyme protein.
[0031] (3) The enzyme protein solution in step (2) is dialyzed to obtain a protein sample solution, passed through a Q Sephrose FF ion-exchange chromatography column, first eluted with Buffer A until no protein flows out, and then gradiently eluted with the eluent Buffer B, Obtain high-purity tyrosinase.
Embodiment 2
[0032] Embodiment 2: Use MS to identify and determine the partial amino acid sequence of purified tyrosinase, design primers according to the amino acid sequence obtained by MS, and clone a part of the tyrosinase coding gene, its main features are:
[0033] The genotype of the upstream primer is: TCCCCCTCGTTCCTGCCCTGGCACCG;
[0034] The genotype of the downstream primer is: GGTGCCGCCGAGGAAGTCGGGCGCCC;
[0035]The PCR amplification conditions of the gene encoding tyrosinase were: pre-denaturation at 94°C for 5 minutes; denaturation at 95°C for 45 seconds, annealing at 69°C for 30 seconds, extension at 72°C for 30 seconds, and 25 cycles of reaction;
[0036] After PCR amplification, use 0.8% agarose gel electrophoresis, Goldview staining, observe the results under ultraviolet light, connect the pMD-18T vector to transform Escherichia coli E.coli JM109 after gel recovery, and send it to Shanghai Bioengineering after screening on Amp plate The company performs sequencing. In NCB...
Embodiment 3
[0037] Embodiment 3: the partial gene sequence that embodiment 2 obtains, design primer, apply TAIL-PCR to amplify known gene fragment two-terminal sequence, its main feature is:
[0038] Use SP1-6 combined with AD primers to amplify the 3' end gene sequence of known gene fragments, and use SP7-12 combined with AD primers to amplify the 5' end gene sequences of known gene fragments;
[0039]
[0040] The first round of TAIL-PCR amplification conditions are: 94°C, 1min→97°C, 5min→{95°C, 30s, 67°C, 1min, 72°C, 2min30s}×5cycles→94°C, 30s, 30°C, 3min, ramp to72°C over 3min, 72°C, 2min30s→{94°C, 30s, 67°C, 1min, 72°C, 2min30s, 94°C, 30s, 67°C, 1min, 72°C, 2min30s, 94°C, 30s, 44°C, 1min, 72℃, 2min30s}×15cycles→store at 12℃;
[0041] The first-round TAIL-PCR product was diluted 40 times as the second-round TAIL-PCR template, and its amplification conditions were:
[0042] 94°C, 1min→{94°C, 30s, 67°C, 1min, 72°C, 2min30s, 94°C, 30s, 67°C, 1min, 72°C, 2min30s, 94°C, 30s, 44°C, 1mi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com