A new closed-type visual nucleic acid detection method coupled with nucleic acid amplification reaction, nucleic acid invasion reaction and nanoparticle color reaction
A technology of nucleic acid invasion reaction and nucleic acid amplification reaction, which is applied in the field of molecular biology, can solve the problems of cross-contamination of amplification products, low sensitivity, and low discrimination, and achieve low-cost, high-sensitivity, and high-specificity effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Example 1 The single-tube closed-tube visual nucleic acid detection method using nucleic acid amplification reaction combined with cascade nucleic acid invasion reaction and nano-gold color detection to detect ultraviolet quantitative genomic DNA.
[0063] The whole genome DNA is used as a template for detection after gradient dilution to investigate the sensitivity of the method of the present invention.
[0064] Reaction conditions:
[0065] The reaction system in this step is composed of 10mM Tris-HCl (pH7.5), 0.05% Tween-20, 0.05% NonidetP40, and 4mM MgCl 2 , 30mM NaCl, 0.5μM upstream primer (sequence: 5'-CCG GGA GTT GGG CGA GTA C-3', Tm value 70.8℃, SEQ ID NO.1), 0.5μM downstream primer (sequence: 5'- GCC CCC AGC AGG TCC C-3', Tm value is 71.8℃, SEQ ID NO.2), 0.2μM upstream probe (sequence is: 5'-CTC TGC CAC CAG CTC CCA-3', Tm value is 68.5℃ , SEQ ID NO.3), 0.2μM downstream probe (the wild-type probe sequence is: 5'-CGC GCC GAGGCG AAC TTG GGC GAG-C 3 -3’, Tm value is 62....
Embodiment 2
[0067] Example 2 A single-tube closed-tube visual nucleic acid detection method using nucleic acid amplification reaction combined with cascade nucleic acid invasion reaction and nanoprobe color detection is used to detect SNP site typing.
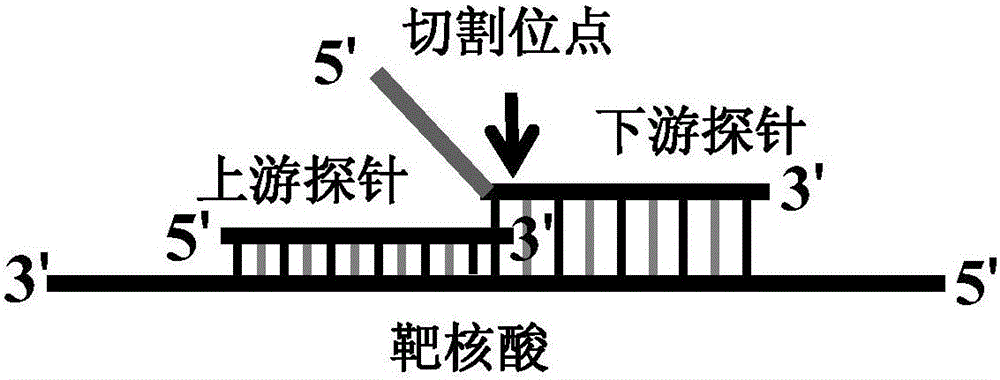
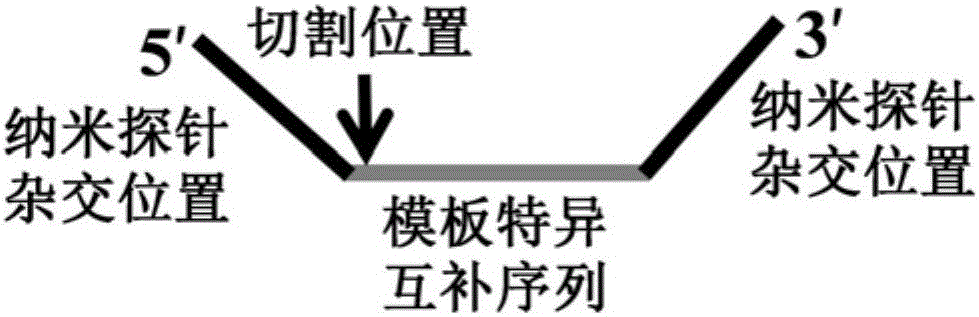
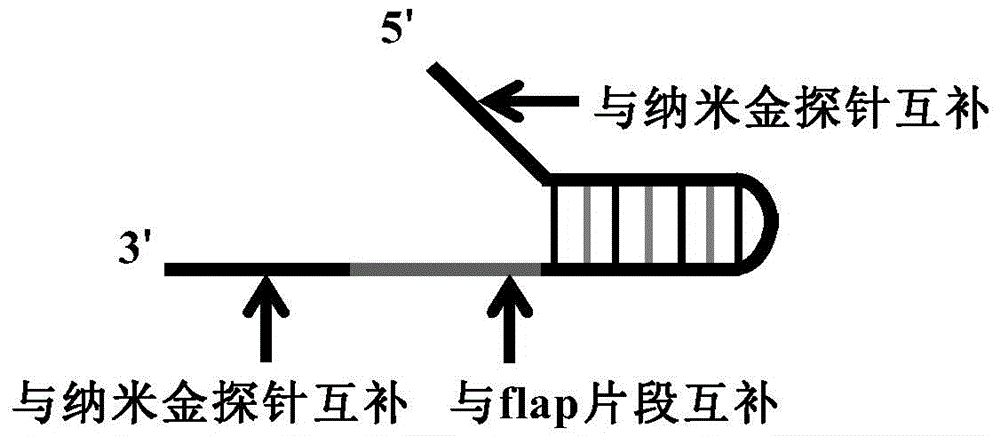
[0068] The detection of SNP site typing using the present invention requires the corresponding downstream probes to be designed for different types of detection sites, and the detection is carried out in two tubes. When the reaction system contains a template that matches the downstream probe, it hybridizes with the nanoprobe When the reaction system does not contain a template that matches the downstream probe, it will aggregate after hybridization with the nanoprobe, thereby realizing the typing of SNP sites.
[0069] Reaction conditions:
[0070] The reaction system in this step is composed of 10mM Tris-HCl (pH7.5), 0.05% Tween-20, 0.05% NonidetP40, and 4mM MgCl 2 , 30mM NaCl, 0.5μM upstream primer (sequence: 5'-CCG GGA GTT GGG CGA GTA C-3', T...
Embodiment 3
[0072] Example 3 The single-tube closed tube of nucleic acid amplification reaction combined with cascade nucleic acid invasion reaction and nano-gold color detection visualizes the effect of different storage time on nano-gold precipitation after nucleic acid detection.
[0073] The whole genome DNA is used as a template for detection, and after the detection reaction is completed, it is placed at room temperature for different times to investigate the precipitation of the nano-gold aggregation reaction in the method of the present invention.
[0074] Reaction conditions:
[0075] The reaction system in this step is composed of 10mM Tris-HCl (pH7.5), 0.05% Tween-20, 0.05% NonidetP40, and 4mM MgCl 2 , 30mM NaCl, 0.5μM upstream primer (sequence: 5'-CCG GGA GTT GGG CGA GTA C-3', Tm value 70.8℃, SEQ ID NO.1), 0.5μM downstream primer (sequence: 5'- GCC CCC AGC AGG TCC C-3', Tm value is 71.8℃, SEQ ID NO.2), 0.2μM upstream probe (sequence is: 5'-CTC TGC CAC CAG CTC CCA-3', Tm value is 68.5...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com