Method for improving protein expression efficiency by employing model fitting and gene modification and application thereof
A technology of genetic modification and protein expression, applied in the fields of plant gene improvement, chemical instruments and methods, and botanical equipment and methods, etc., can solve the problems of reducing the persistence and efficiency of expression, and achieve the improvement of expression efficiency, increase expression efficiency, and high efficiency. The effect of versatility and feasibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] mRNA secondary structure fitting
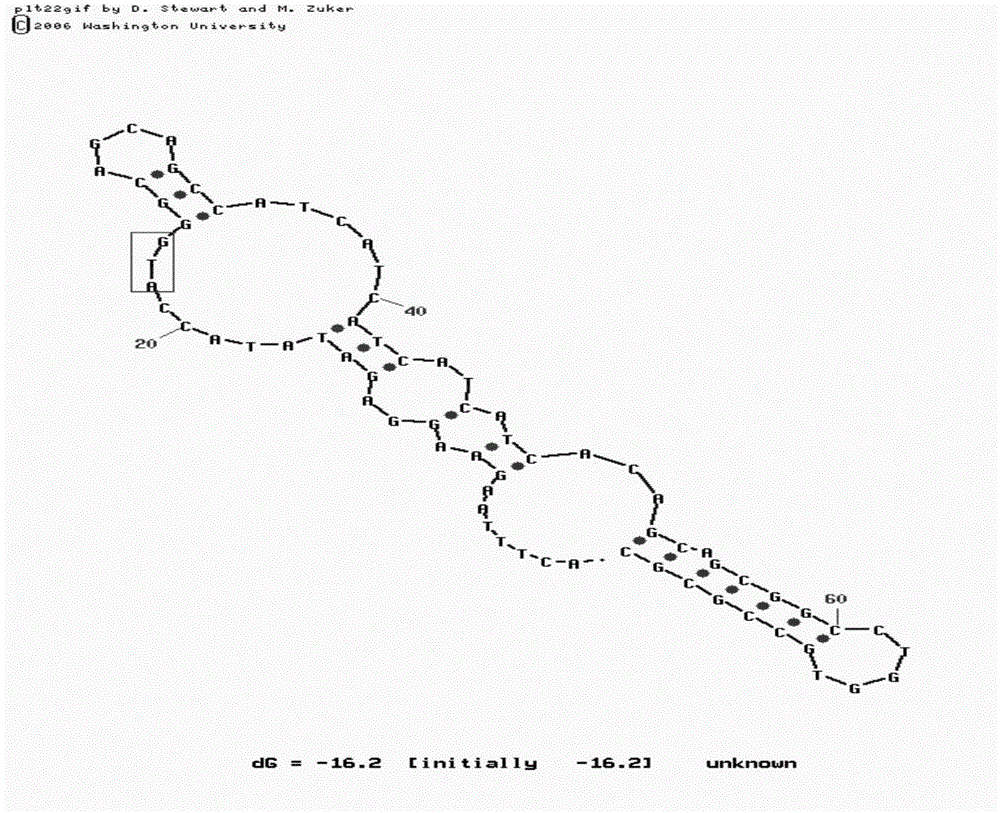
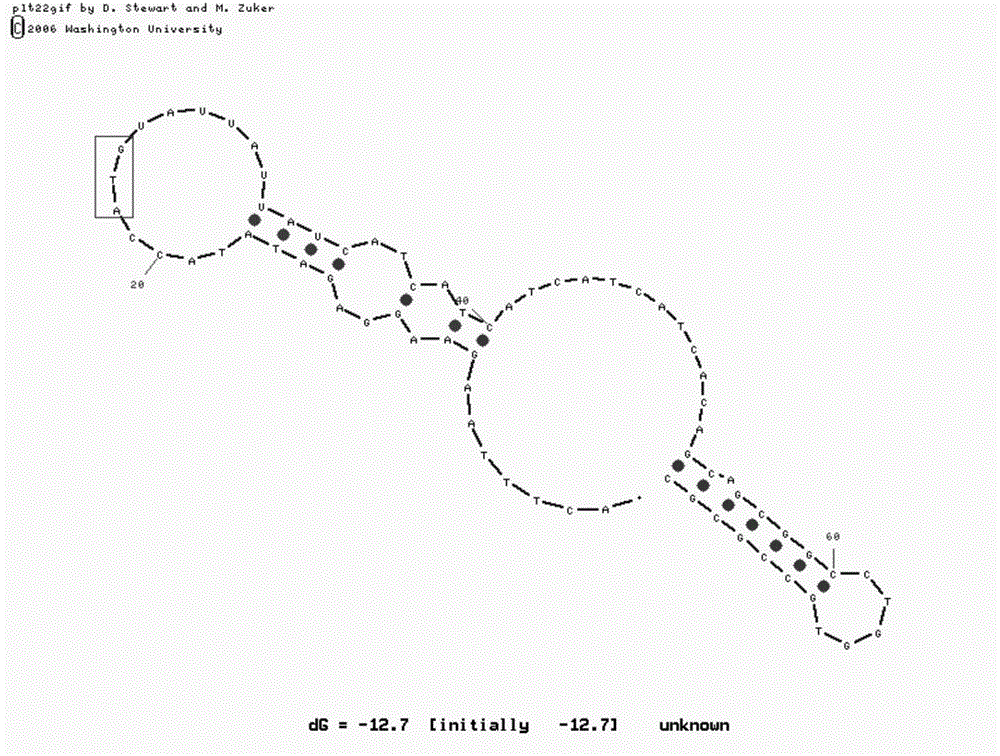
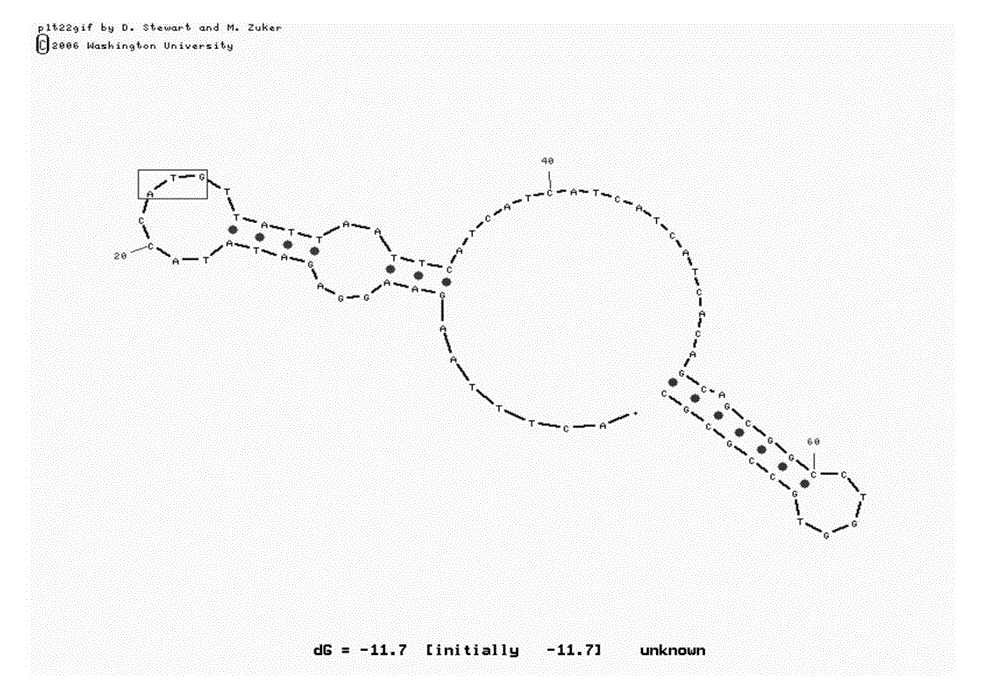
[0031] In the M-fold program, input the mRNA sequence of L11. Since the sequence before the start codon, including the SD sequence, etc., can form hydrogen bonds and secondary structures with the sequence after the start codon, and can affect the translation speed of the protein, therefore, enter about 20 nucleotide sequences Perform secondary structure fitting; at the same time, input about 50 nucleotide sequences after the start codon to ensure that the mRNA has enough length to fold after the start codon.
[0032] After fitting with M-fold, it was found that the mRNA of wild-type L11 had a hairpin structure immediately after the start codon ( figure 1 ). Therefore, when the ribosome is translating a protein, it is very likely that reading the mRNA sequence at this position will be blocked, reducing the speed of translating the protein, and ultimately leading to a decrease in the yield of the recombinant protein. At the same time,...
Embodiment 2
[0034] L11 plasmid purification
[0035] A single colony of BL21(DE3)pLysS L11 was inoculated into 20 ml of LB medium containing 10 μg / ml ampicillin (Sigma-Aldrich), and the flask was incubated overnight at 37°C with shaking. The next day, 5ml of saturated BL21(DE3)pLysS L11 cells were transferred to 500ml of terrific broth (Invitrogen) with 10 μg / ml ampicillin, and the flask was incubated overnight at 37°C with shaking. Cells were harvested by centrifugation at 5,500 rpm for 10 minutes with a GS-3 rotor of a Beckman centrifuge. Cells were snap frozen in liquid nitrogen and stored at -80°C.
[0036] Cells were lysed in 20 ml of a solution containing 1 mg / ml lysozyme (Sigma-Aldrich). After the solution was homogeneous, 40 ml of 1% SDS in 0.2N NaOH solution was added to precipitate DNA of chromosome and plasmid at the same time. The solution was kept on ice for 10 minutes and 20 ml of 5M potassium acetate solution (pH 4.8) was added to precipitate the chromosomal DNA while th...
Embodiment 3
[0039] Design of primers for site-directed mutagenesis of L11 plasmid
[0040] The Stratagene QuikChange site-directed mutagenesis kit for vectors was used to construct multiple mutants. Primer design is based on the following principles: 1. The two primers must contain the same mutated sequence on the opposite plasmid strand; 2. The length of the primer should be between 25-45 bases, and the melting temperature (Tm) ≥ 78 °C ; 3. The position of the desired mutation (deletion or insertion) should be in the middle of the primer, and there are 10 to 15 bases with correct sequence on both sides; 4. The minimum cytosine and guanine (GC) content of the primer is 40%, And terminate at one or more C or G bases.
[0041] T m =81.5+0.41(%GC)-675 / N-mismatch % (reaction formula 1)
[0042] T m =81.5+0.41(%GC)-675 / N (for insertion or deletion) (reaction formula 2)
[0043] N is the length of bases in the primer, GC percentage and mismatch percentage are integers.
[0044] The design...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com