Kit for detecting klebsiella pneumoniae
A Klebsiella and kit technology, applied in the field of PCR detection of strains, can solve the problems of low detection rate, low accuracy, poor repeatability, etc., and achieve accurate identification, increase the positive rate, and shorten the detection cycle. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Embodiment 1 Fluorescence quantitative PCR detects the establishment of the method for Klebsiella pneumoniae
[0041] 1. Primer and probe design
[0042] For the conserved region of the phoE gene of Klebsiella pneumoniae, this embodiment designs 3 pairs of primers and the probes used in conjunction with it. The 3 pairs of primers are all located on the phoE gene, and their nucleotide sequences are as follows:
[0043] (1)
[0044] Upstream primer: 5'-GATGGGCTTTGTGGCTTCAA-3'
[0045] Downstream primer: 5'-CCAGCTTGTTCGCGTTCTTAT-3'
[0046] Probe: 5'-AGCGACGCAGGCAGCGGAA-3'
[0047] (2)
[0048] Upstream primer: 5'-CGATATGTTCCCGGAATTCG-3'
[0049] Downstream primer: 5'-GCTGGCGCGCTTGGT-3'
[0050] Probe: 5'-CGGCGACTCATCCGCTCAGACC-3'
[0051] (3)
[0052] Upstream primer: 5'-GGCCGTTGGGAATCTGAATT-3'
[0053] Downstream primer: 5'-CGAACGCCAGACGAGTCTTT-3'
[0054] Probe: 5'-AACCGAGAGCGACTCCAGCCAGC-3'
[0055] The 5' end of the Taqman probe nucleotide sequence is marked ...
Embodiment 2
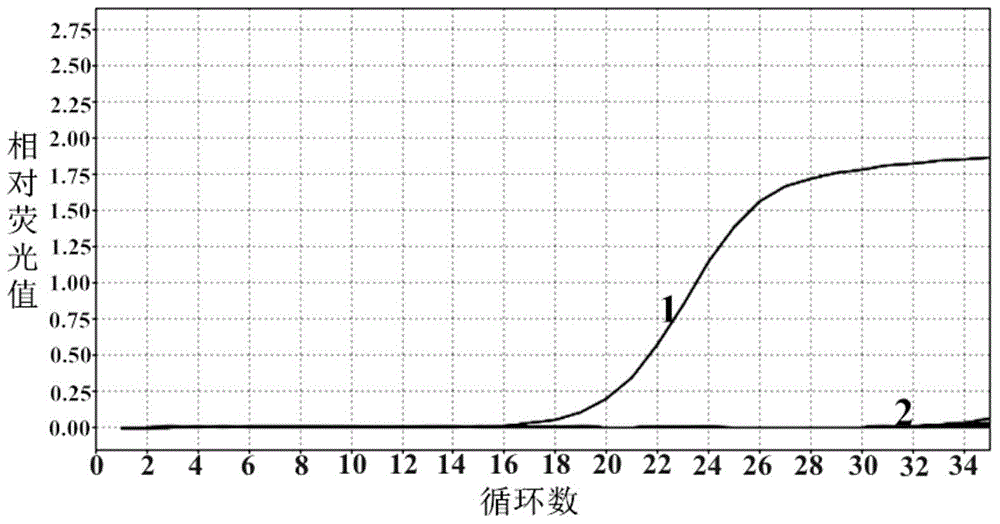
[0069] Embodiment 2 Fluorescence quantitative PCR detects the specificity verification of Klebsiella pneumoniae method
[0070] Klebsiella pneumoniae, Enterococcus faecium, Enterococcus faecalis, Staphylococcus aureus, Staphylococcus epidermidis, Escherichia coli, Acinetobacter baumannii, Enterobacter cloacae, Staphylococcus hemolyticus in LB medium for 37 days Cultivate overnight at ℃, and extract genomic DNA from the bacterial solution. The extracted genomic DNA of the above-mentioned bacteria was used as a template, and sterilized deionized water was used as a negative control, and the Taqman probe method fluorescent quantitative PCR reaction was carried out. The result is as figure 2 , Klebsiella pneumoniae template amplification curve is S line, Ct value is 16, Enterococcus faecium, Enterococcus faecalis, Staphylococcus aureus, Staphylococcus epidermidis, Escherichia coli, Acinetobacter baumannii, Enterobacter cloacae , Staphylococcus hemolyticus and the negative contr...
Embodiment 3
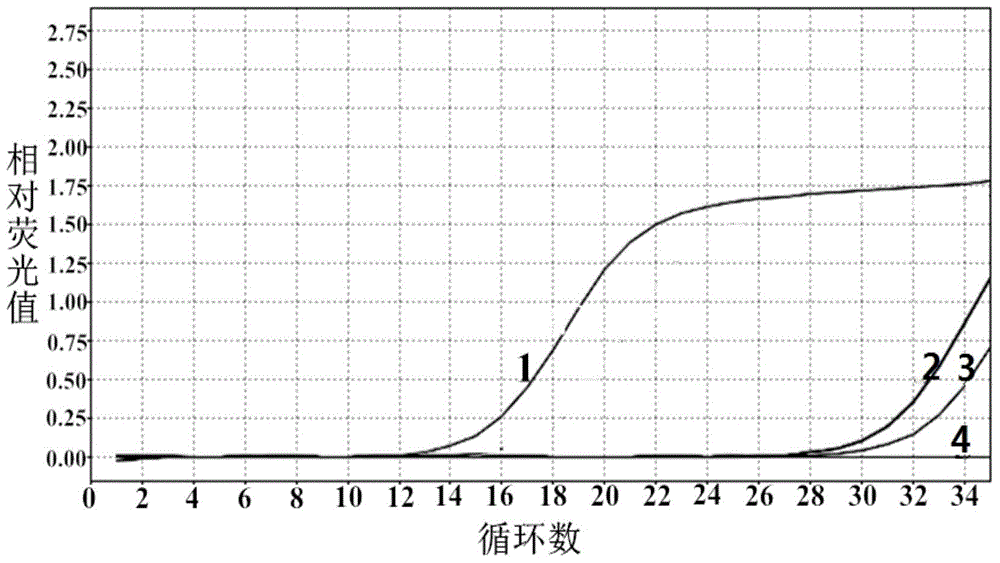
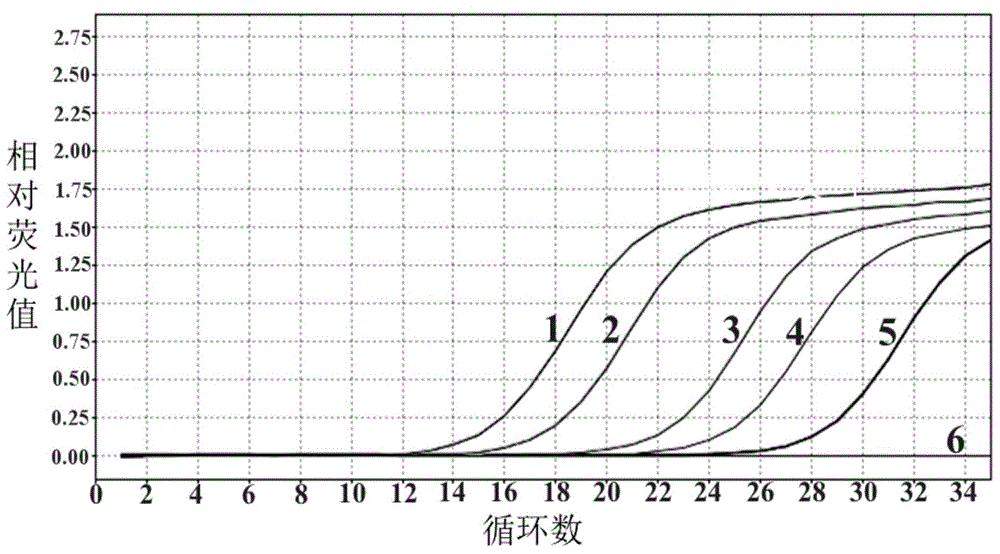
[0071] Example 3 Sensitivity verification of the method for detecting Klebsiella pneumoniae by fluorescent quantitative PCR
[0072] Klebsiella pneumoniae was cultured overnight at 37°C in LB medium, counted colonies, and then the concentration was 1 × 10 6 CFU / ml bacterial solution was diluted 10 times to 1×10 2 CFU / ml. The genomic DNA of each concentration of bacterial liquid was extracted as a template, and sterilized deionized water was used as a negative control, and the Taqman probe method was used for fluorescent quantitative PCR reaction. The result is as image 3 display, 1 represents 1×10 6 Template for CFU / ml bacterial solution, 2 represents 1×10 5 Template for CFU / ml bacterial solution, 3 represents 1×10 4 Template for CFU / ml bacterial solution, 4 represents 1×10 3 Template for CFU / ml bacterial solution, 5 represents 1×10 2 CFU / ml bacterial liquid template, in a 20μl reaction system under optimal reaction conditions, the minimum detection limit is 1×10 2 Th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com