SSR primer group and method for malt variety identification by virtue of primer group
A variety identification and primer group technology, applied in biochemical equipment and methods, microbial measurement/testing, recombinant DNA technology, etc., can solve problems affecting saccharification, filtration and brewing performance, secondary germination, and difficulty in separation , to achieve the effect of preventing malt adulteration, maintaining brand image, and increasing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
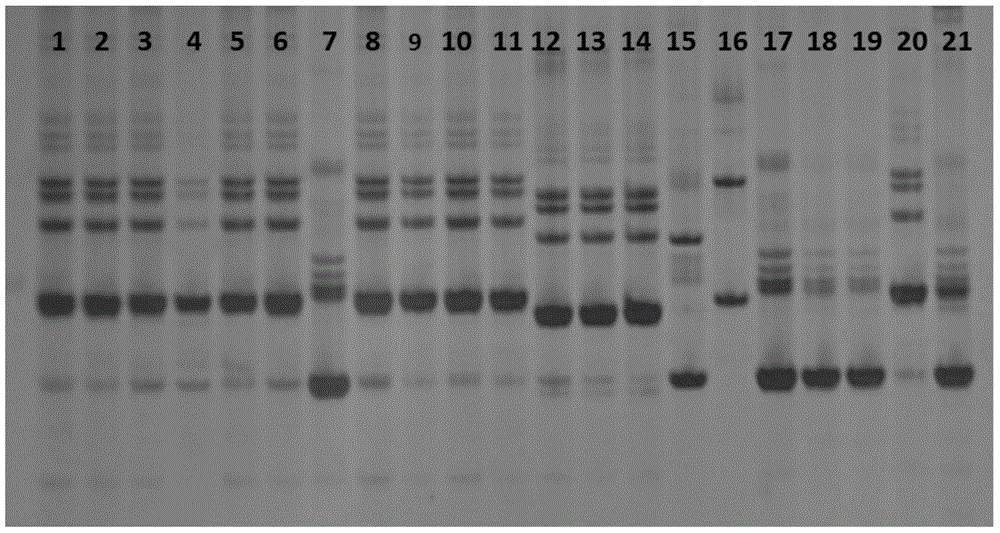
[0035] Select 21 barley samples from different origins, germinate and dry them into malt.
[0036] Table 1.21 Malt samples
[0037]
[0038] (1) Extract the DNA of the test variety sample
[0039] Use CTAB method to extract DNA from samples of tested varieties: Take a single malt sample in a 1.5ml centrifuge tube, about 25-45mg, add 7-8mm steel balls, and use Thmorgan CK1000D high-throughput tissue grinder to crush. Add 600ul of CTAB extraction buffer preheated at 65°C to the sample, and vortex to mix the lysed sample. Add 4ul β-mercaptoethanol and 1% PVP (polyvinylpyrrolidone) to prevent polyphenols from oxidative browning and DNA degradation, effectively remove polyphenols and polysaccharides, and vortex to mix. Water bath at 65°C for 15 minutes, shaking gently during mixing. Add 600ul chloroform / isoamyl alcohol (24:1), vortex and mix vigorously for 20s to form an emulsion. Centrifuge at 10,000 rpm for 10 minutes at room temperature, pipette 300ul of the supernatant into a new ...
Embodiment 2
[0052] The sample includes 2 test malt varieties, one is the control variety Copeland with 2 grains, and the other is the malt variety A to be identified with 6 grains, to determine whether the variety A is Copeland.
[0053] (1) Prepare malt DNA for the sample, use 6 pairs of SSR primers for PCR amplification, polyacrylamide gel electrophoresis, silver staining, and map analysis. The specific implementation steps are as in Example 1 (1)-(4).
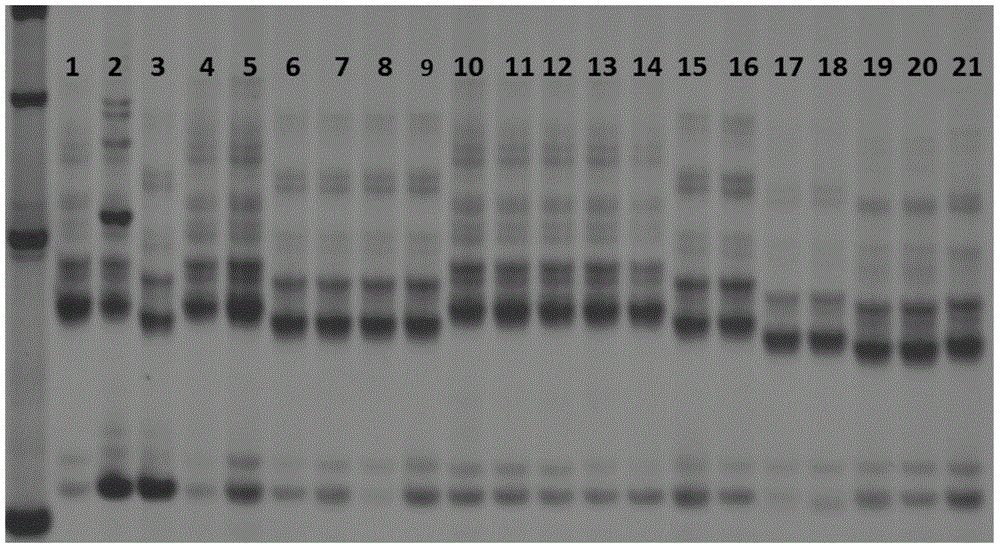
[0054] (2) Result analysis: by Figure 7 It can be seen that lanes 1-8 are the first pair of primers scssr10148, lanes 1 and 2 are the control species Copeland, lanes 3-8 are the species to be identified, and the electrophoretic bands in lanes 3, 5, 6, and 7 are compared with lanes 1, 2 The electrophoresis bands of the samples are completely the same, but the electrophoresis bands in lanes 4 and 8 are different from the control sample Copeland, so it can be determined that lanes 4 and 8 are other varieties. Lanes 9-16 are the second pair of ...
Embodiment 3
[0056] The samples included 2 test malt varieties, one was the control variety Metcalfe with 2 grains, and the other was the unidentified malt variety B with 6 grains.
[0057] (1) Prepare malt DNA for the sample, use 6 pairs of SSR primers to perform PCR amplification, polyacrylamide gel electrophoresis, silver staining, and map analysis. The specific implementation steps are as the steps (1)-(4) of Example 1.
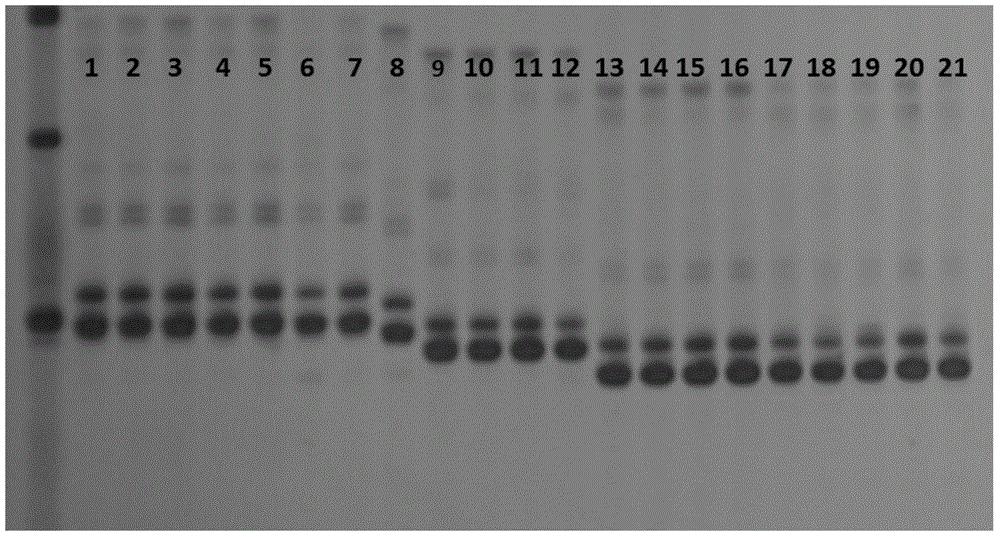
[0058] (2) Result analysis: by Figure 8 It can be seen that lanes 1-8 are the first pair of primers scssr10148, lanes 1 and 2 are the control variety Metcalfe, and lanes 3-8 are the varieties to be identified. The electrophoresis bands in lanes 4-8 are the same as those of the control samples in lanes 1 and 2. The bands are completely the same, but the electrophoretic bands in lane 3 are different from the control sample Metcalfe, which can confirm that lane 3 is of other varieties. Lanes 9-16 are the second pair of primers Bmac0030, lanes 9, 10 are the control varieties ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com